The CRISPR/Cas9 System and its Applications
Authored by Debbie King, Contributor, Cell Culture Dish
Evolution of Gene Editing
Genome editing in vitro has been studied since the 1970’s with the discovery that exogenous DNA could be taken up by yeast or bacteria and randomly integrated into the genome. Subsequently, targeted integration of DNA into the yeast (Saccharomyces cerevisiae) genome was demonstrated (Scherer et al., 1979), opening the doors for genetic manipulation. Since then, progress in applications of genome editing (i.e. Gene Therapy) has been limited due to lack of efficient, inexpensive, and accurate methodologies.
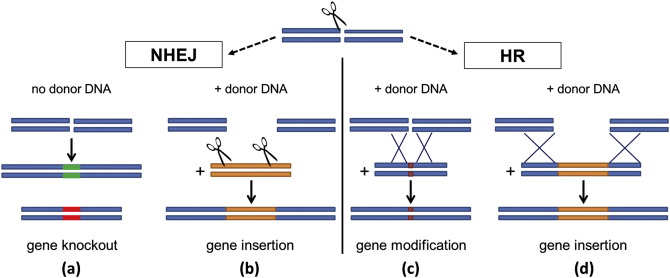
In general, a gene of interest is targeted to modify its function, and a nuclease (an enzyme that cleaves nucleic acids) cuts that gene’s DNA sequence, breaking the structure of the DNA. After cleavage, a new gene can be inserted or a change to the existing sequence can be made or deletion of a specific stretch of genomic DNA can be made (Figure 1).
Until 2013, zinc finger nucleases (ZFNs, Kim et al 1996) and transcription activator-like effector nucleases (TALENs, Christian et al, 2010) were the dominant technologies for gene editing. Both systems utilize artificial fusion proteins composed of an engineered DNA-binding domain fused to FokI nuclease. There are drawbacks to both systems: 1) they require costly and time-consuming protein synthesis for each unique DNA target sequence and 2) they cannot access certain DNA sites. For example, ZF and TALENs cannot bind to methylated DNA sites therefore making gene editing at these sites impossible with these systems and limiting gene targets.
CRISPR/Cas9 System
The latest ground-breaking technology for genome editing called Clustered Regularly Interspaced Short Palindromic Repeat CRISPR/Cas9 system is based on RNA-guided nucleases, which has great potential due to their simplicity, efficiency and versatility. The most widely adopted system is the type II CRISPR/Cas9 system from Streptococcus pyogenes (Jinek et al., 2012).
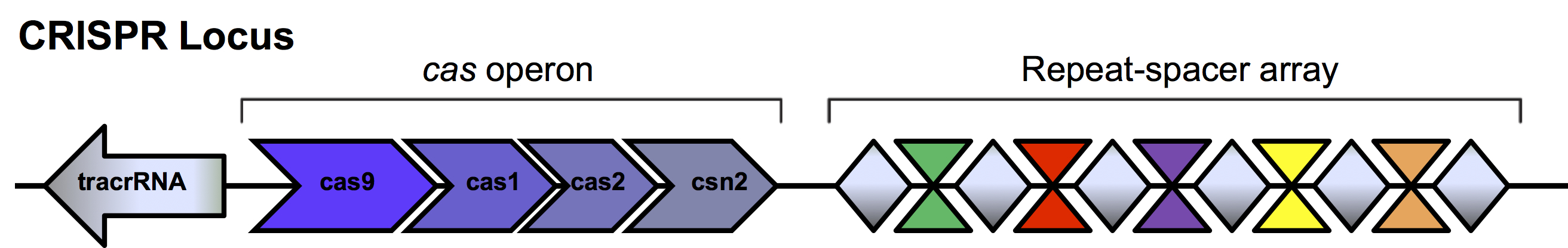
The presence of these repeats was first described in Escheria coli in 1987 (Ishino et al., 1987). Unique DNA sequences located between the repeats called spacers were identified but their function was not elucidated until 2005 when homology was found to viral and plasmid sequences (Bolotin et al., 2005, Mojica et al., 2005, Pourcel et al., 2005). It was suggested these spacers were acquired as a defense mechanism in microbes to invading phages and plasmids. When foreign DNA enters the bacterial cell, enzymes encoded by the bacteria cut the invading DNA into small fragments. One or more of these unique spacers is inserted into the bacterial genome between the short repeat sequences within the repeat-spacer array in the CRISPR loci (Figure 2). This newly altered CRISPR locus is passed on to subsequent generations as the bacteria divides. This so-called “adaptive immunity” allows bacteria to retain a genetic memory of viruses and plasmids it has encountered.
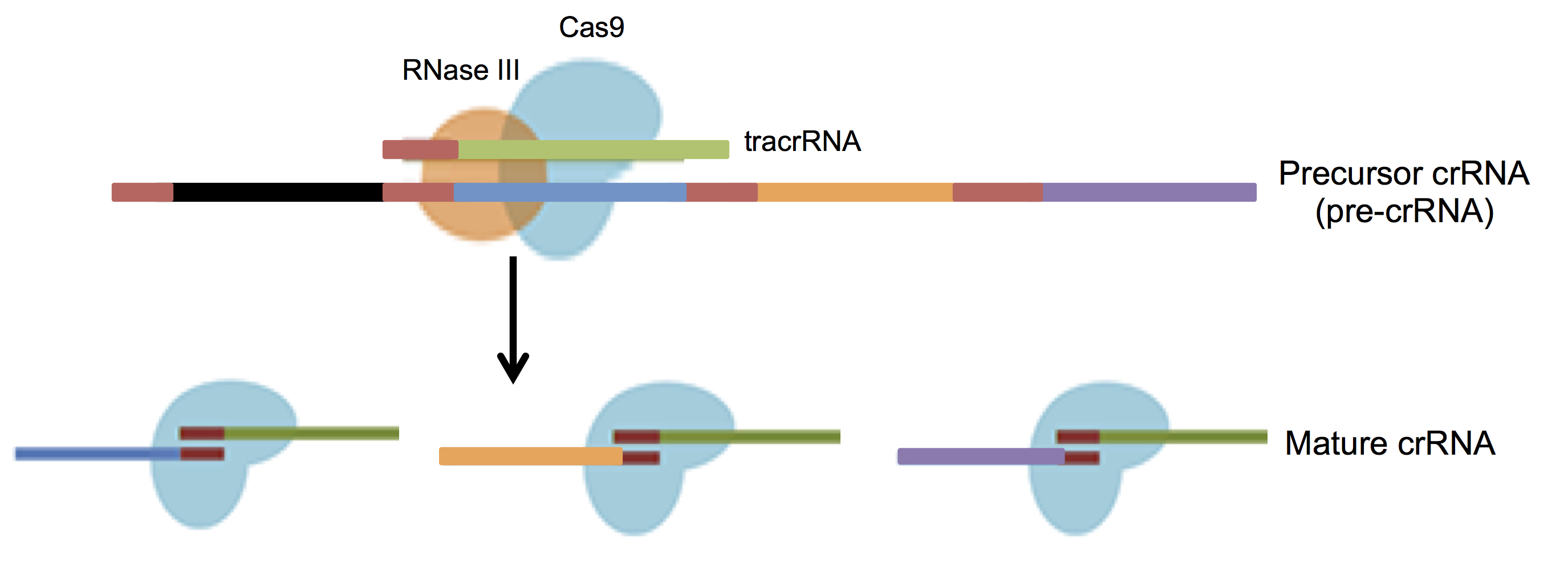
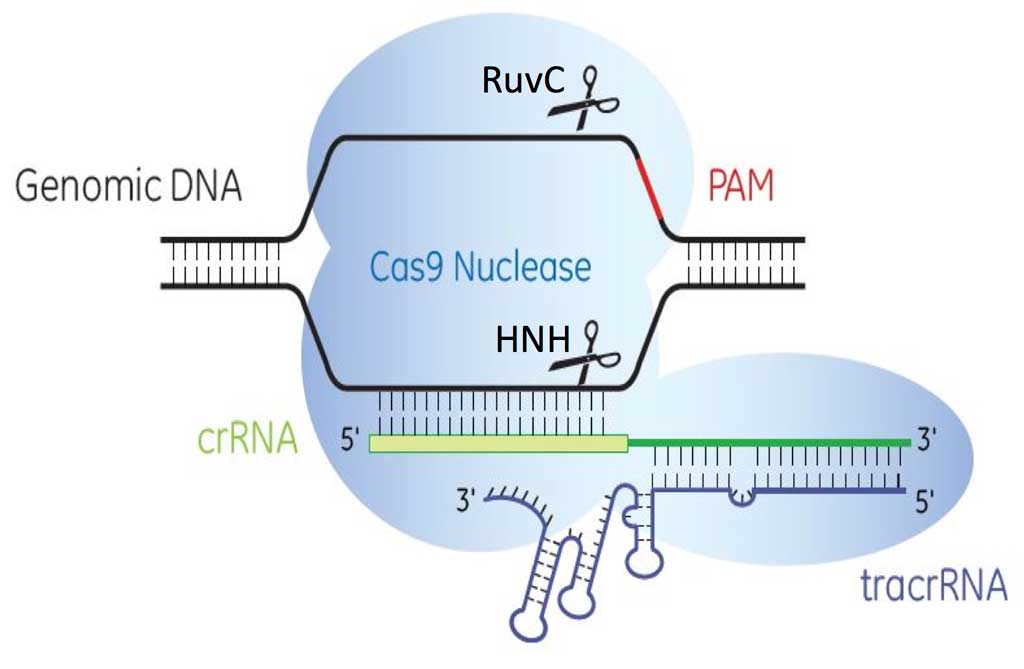
Transcription of the CRISPR loci occurs when the bacteria encounters invading DNA, which generates a long precursor CRISPR RNA (pre-crRNA). The pre-crRNA is processed into mature crRNAs by host enzymes when cleaved upstream of each spacer generating many small crRNAs of ~40 nucleotides in length. A small RNA sequence upstream of the S. pyogenes CRISPR loci transcribes a trans-activating CRISPR RNA (tracrRNA, Barrangou et al., 2007) that combines with mature crRNA to activate and guide Cas9 nuclease to the appropriate DNA target (Figure 3).
There are three identified CRISPR-Cas systems: I, II and III (Markarova et al., 2011), each having distinct mechanisms to achieve acquired immunity. Types I and II CRISPR/Cas9 systems require the presence of protospacer adjacent motifs (PAMs), which are short sequences, 5’-NGG-3’, downstream of the 20 nucleotide crRNA-targeted sequence for Cas9 nuclease binding. Cas9 nuclease has two domains, the HNH (an endonuclease domain named for characteristic histidine and asparagine residues) and RuvC (an endonuclease domain named for an E. coli protein involved in DNA repair), responsible for cleaving both strands of DNA with the HNH domain cleaving the strand complementary to the 20nt crRNA sequence. The presence of three components is needed to effectively target and cleave DNA: 1) crRNA, 2) tracrRNA and 3) Cas9 endonuclease (Figure 4).
This prokaryotic system showed functionality when introduced into eukaryotic cells (Cho et al., 2013; Cong et al., 2013; Hwang et al., 2013; Jinek et al., 2013; Mali et al., 2013). For the purposes of gene editing in eukaryotic cells, the two-component system of crRNA:tracrRNA used to recruit Cas9 was simplified into one chimeric RNA molecule called the single guide RNA (sgRNA, Jinek et al., 2012). The sgRNA retains two critical features: the 20-nucleotide sequence at the 5′ end that determines the DNA target site by Watson-Crick base pairing, and the double-stranded structure at the 3′ side of the guide sequence that binds to Cas9 (Figure 5).
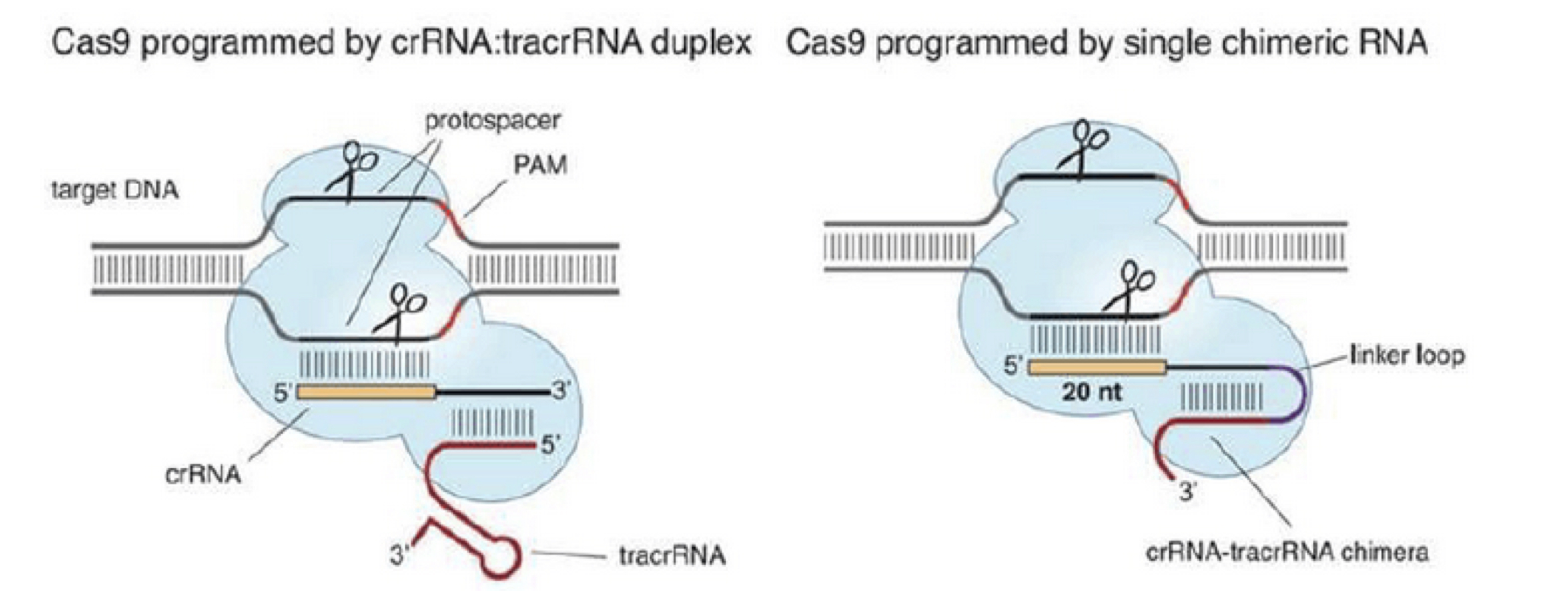
This modification enables very specific RNA-targeted DNA cleavage simply by altering the sgRNA sequence which significantly reduces the time, cost and technical difficulty of gene editing.
CRISPR/Cas9 Applications
Basic Science
To date, the CRISPR/Cas9 gene editing tool appears to work in nearly every organism, from Caenorhabditis elegans to monkeys, and in every cell type: kidney, heart and those, like T-cells, that researchers had previously found difficult to modify.
The CRISPR interference technique was first reported in 2013 (Qi et al., 2013). The ability of Cas9 nuclease to bind to specific DNA sites with high fidelity makes it attractive for study of gene function. Cas9 endonuclease activity can be deactivated to prevent DNA cleavage at the target site (denoted as dead Cas9 or dCas9) and activation or repression of target genes can be achieved. dCas9 targeted to the promoter region of a specific gene can repress transcription through steric hindrance, whereas, activation can be achieved through fusion of an activator protein to the deactivated enzyme. Once bound to the promoter region, it can stimulate transcription of the targeted gene. Specifically, fusion of dCas9 to effector domains including VP64 or KRAB allowed stable and efficient transcriptional activation or repression, respectively, in human and yeast cells (Gilbert et al., 2013).
Imaging in live cells can also be accomplished when dCas9 is tagged with fluorescent proteins or small molecules (Chen et al., 2013).
Agriculture
Some of the earliest papers using CRISPR/Cas9 for gene editing focused on crops important for agriculture such as rice and wheat. It was realized early on that this system could be utilized to specifically alter the DNA of these crop species to improve traits like disease and drought tolerance. Conventional breeding of desirable traits relies on existing natural genetic variation and extensive back-crossing programs. Wang et al. (2014) used both TALEN and CRISPR/Cas9 technologies to target and successfully knock out the genes of the mildew-resistance locus (MLO) in wheat to generate plants resistant to powdery mildew disease.
Big Agriculture like DuPont has announced its intention to use CRISPR–Cas9 technology to engineer crops in partnership with genome-editing firm Caribou Biosciences of Berkeley, California.
A team led by a researcher at the University of California, Riverside (Swartz et al., 2016) has adapted the CRISPR/Cas9 system for use in oleaginous (oil-producing) yeast Yarrowia lipolytica. This yeast strain has been difficult to genetically manipulate with previously available technology. The CRISPR/Cas9 system will allow Y. lipolytica to be directed to produce long chain hydrocarbons – used for polymers, adhesives, coatings and fragrances – most often obtained from non-renewable sources (petroleum) because they are difficult to synthesizeand also to produce precursors for biofuels relieving the dependence on edible plant oils.
Disease Modeling
Disease modeling using animals has been a hurdle for study of many types of human diseases due to the difficulty in generating genetically modified animals that accurately recapitulate human pathology. The process was inefficient and inaccurate or simply impossible to do. CRISPR-Cas9 has allowed for generation of transgenic animals like rats, monkeys and other animals which are more suitable for human disease modeling than mice and thus permit better drug-development tests.
Neuroscientists have long awaited transgenic monkeys specifically for disorders involving the brain like autism, schizophrenia and Alzheimer’s disease which cannot be fully recapitulated in mice since they do not have the complex cognitive abilities of primates. For example, scientist Guoping Feng at MIT is using the method to engineer marmosets that have a disrupted SHANK3 gene, which is a candidate autism gene (Shen, 2013). CRISPR-Cas9 was used to alter the DNA of the single-cell monkey embryo causing the disrupted gene to be present in every cell of the primate. This will better allow study of the gene’s role in autism and allow identification of novel therapeutics.
Gene Therapy
In 2014, CRISPR-Cas9 was used in mice to correct a mutation associated with a human metabolic disease called tyrosinaemia (Yin et al., 2014). It was the first use of CRISPR/Cas9 to fix a disease-causing mutation in an adult animal and an important step towards using the technology for Gene Therapy in humans.
HIV-1 (Human Immunodeficiency Virus) is a virus able to integrate its genome into the host cell DNA during infection. HIV-1 primarily infects CD4+ T cells and leads to AIDS (acquired immunodeficiency syndrome) if untreated. The viral DNA is inserted into the host cell genome by integrases which use long terminal repeat (LTR) sequences on the viral genome (Ebina et al., 2013). AIDS can be controlled by anti-retroviral therapy (targeting the reverse-transcription step in the HIV replication process). However, the latent pool of HIV-1 DNA in the host cell remains unaffected by this therapy so the disease cannot be cured. CRISPR-Cas9 was used to edit out copies of latent HIV-1 DNA from CD4+ T cell genomes in human lab cultures (2D10 CD4+ T-cell line) by targeting the 5’ and 3’ LTRs of the viral genome (Kaminski et al., 2016). When these cells were later exposed to the virus, they were protected from re-infection by HIV-1 virus. These findings are a stepping stone to translating such a system to T-cell-directed HIV-1 therapies.
Big Pharma has raced ahead with an eye towards clinical therapies using CRISPR/Cas9 system. Editas Medicine (NASDAQ: EDIT) is slated to begin clinical trials in 2017 to treat a rare form of blindness, Leber Congenital Amaurosis (LCA10). The CRISPR/Cas9 system will be used to correct a mutation in a gene encoding a centrosomal protein important in centrosome and cilia development, CEP290. In addition, Intellia Therapeutics recently filed for IPO (April 2016) having cemented a deal with Regeneron (NASDAQ: REGN) to develop Gene Therapy for transthyretin amyloidosis (ATTR) by delivering CRISPR/Cas9 to edit the genes of a patient’s liver cells using lipid nanoparticles. ATTR is one of three specific liver diseases Intellia says it is targeting (Lash, 2016).
CRISPR Considerations
Jennifer Doudna, a pioneer of the CRISPR-Cas9 gene editing system, described during a TED talk work by a postdoctoral student to create a lung cancer mouse model using virally packaged CRISPR/Cas9 in an inhalable virus (Doudna, TED Talk, 2015). While this research was exciting, if an error occurred encoding the sgRNA with specificity for the mouse genome, the virus carrying the cancer-causing mutations could potentially be introduce into human lungs. CRISPR/Cas9 has made gene editing more accessible than ever but caution must be exercised with such a powerful technique.
Another hurdle is the off-site targeting effects during gene editing, particularly for scientists who want to treat human disease due to the risk of tumor formation caused by unintended mutations. This refers to the tolerance of Cas9 to mismatches in the sgRNA sequence and is dependent on the number, position, and distribution of mismatches throughout the sequence. Many studies have been done to examine and address the factors contributing to off-site target mutagenesis.
There are also ethical concerns that CRISPR/Cas9 could be used to alter the DNA in human embryos before the ramifications are truly considered. For such therapies to be viable in the clinic, the gene editing efficiencies need to be near 100% and chances of off-target mutagenesis non-existent. In 2015, Chinese scientists (Liang et al., 2015) attempted to modify the gene HBB encoding the human β-globin protein, responsible for β-thalassaemia a potentially fatal blood disorder, using non-viable human embryos obtained from local fertility clinics. While the authors made clear none of the embryos used could result in a live birth, this research raised some serious ethical questions about editing the human germ line, particularly for clinical applications. On February 1, 2016, scientists in London have been granted permission to edit the genomes of human embryos for research. This approval by the UK Human Fertilization and Embryology Authority (HFEA) represents the world’s first endorsement of such research by a national regulatory authority (Callaway, 2016).
CRISPR Final Remarks
CRISPR/Cas9 gene editing technology is being touted as one of the biggest biotechnology breakthroughs of the century with Jennifer Doudna of the University of California, Berkeley, and Emmanuelle Charpentier of the Helmholtz Centre for Infection Research in Braunschweig, Germany, receiving public accolades for its discovery (winners of Science’s 2015 Breakthrough of the Year). There are even rumors of the Nobel Prize in Chemistry.
However, this triumph in science is marred by controversy over who owns the CRISPR/Cas9 technology. The ongoing patent battle between Doudna/Charpentier (UC Berkeley) and Feng Zhang (Broad Institute, MIT) could be worth millions as it will influence which commercial entities can use this technology. In all likelihood, it will take years to settle this dispute.
Regardless, CRISPR/Cas9 has been revolutionary facilitating a wealth of research unmatched in over three decades since the inception of genome editing. It has opened up gene editing to the broad scientific community and allowed what used to take years to do to be achieved in a matter of weeks. Recent research has also propelled CRISPR/Cas9 system beyond just gene editing tool allowing study of DNA epigenetics and understanding what non-protein coding segments of our genome encodes. Its potential for research, human medicine and agriculture will likely only be limited by our imagination.
References
Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007; 315:1709–12.
Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 2005; 151:2551–61.
Callaway E. UK scientists gain licence to edit genes in human embryos. Nature News, February 1, 2016.
Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, Huang B. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 2013; 155: 1479–1491.
Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA guided endonuclease. Nat Biotechnol 2013; 31:230–2.
Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 2010; 186:757–61.
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013; 339:819–23.
Doudna J. “How CRISPR lets us edit our DNA”. TED. Sept 2015.
Ebina H, Misawa N, Kanemura Y, and Koyanagi Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci Rep. 2013; 3: 2510.
Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013; 154: 442–451.
Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 2013; 31:227–9.
Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol 1987; 169: 5429–33.
Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife 2013; 2:e00471.
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual- RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012; 337: 816–21.
Kaminski R, Chen Y, Fischer T, Tedaldi E, Napoli A, Zhang Y, Karn J, Hu W, Khalili K. Elimination of HIV-1 Genomes from Human T-lymphoid Cells by CRISPR/Cas9 Gene Editing. Scientific Reports, 2016; 6:22555.
Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci U S A 1996; 93:1156–60.
Lash A. “CRISPR Developer Intellia Deals With Regeneron, Jumps Into IPO Queue”. www.xconomy.com April 11, 2016.
Liang P, Xu Y, Zhang X, Ding C, Huang R, Zhang Z, Lv J, Xie X, Chen Y, Li Y, Sun Y, Bai Y, Songyang Z, Ma W, Zhou C, and Huang J. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell. 2015 May; 6(5): 363–372.
Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science 2013; 339:823–6.
Makarova K, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf Y, Yakunin A, van der Oost J and Koonin EV. Evolution and classification of the CRISPR–Cas systems. Nature Reviews Microbiology 2011; 9: 467-477.
Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derived from foreign genetic elements. J Mol Evol 2005; 60: 174–82.
Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 2005; 151: 653–63.
Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013; 152: 1173–1183.
Scherer S and Davis RW. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc. Natl. Acad. Sci. U.S.A., 1979; 76, 4951-55.
Shen H. Precision gene editing paves way for transgenic monkeys. Nature News, November 6, 2013.
Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, et al. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol 2014; 32: 947–51.
Yin H, Xue W, Chen S, Bogorad RL, Benedetti E, Grompe M, Koteliansky V, Sharp PA, Jacks T, Anderson DG. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol. 2014; 32, 551–553.