The Human Protein Atlas
The completion of the Human Genome Project in 2003 to successfully sequence all 20,000 protein-coding genes (proteome) was a monumental scientific achievement, but this was really just the beginning. The next step was to examine the proteome to begin understanding the function of all these proteins.
The Human Protein Atlas (HPA) project was initiated in 2003 with the goal to systematically map the expression of these proteins using antibody-based proteomics. The program is based primarily in Sweden with funding from the non-profit organization Knut and Alice Wallenberg Foundation (KAW). Antibodies, produced in-house and commercially purchased, are first validated by the researchers and then used to characterize each protein. The data is available through an open-access interactive database: www.proteinatlas.org (the first version was launched in 2005). It contains millions of high-resolution images showing the spatial localization of protein expression in 76 different cell types within 44 different normal human tissues (Tissue Atlas) and 20 different cancer types (Cancer Atlas).
Recently, at the American Society for Cell Biology (ASCB, December 2016), a new Cell Atlas was launched in an updated version of the HPA (version 16) during a well-attended Tech Talk. It features:
- high-resolution, multi-color images of immunofluorescent-stained cells
- subcellular spatial localization of proteins in 56 cell lines representative of cell populations in different organs of the human body
- mRNA expression of all human genes characterized using deep RNA-sequencing
- > 12,000 genes with antibodies available mapped to 30 different cellular structures (i.e. nucleus, cytosol, mitochondria, etc.)
Pinpointing the subcellular localization of a protein is critical to understanding their function. As they localize to their intended micro-environment (i.e. organelles, vesicles or macro-molecular complexes), they will encounter other proteins or RNA in suitable biochemical conditions to accomplish their molecular function (Breckles, 2016). This comprehensive atlas provides a much-needed tool for more sophisticated study of protein function by researchers all over the world.
Below are some samples images from the Cell Atlas:

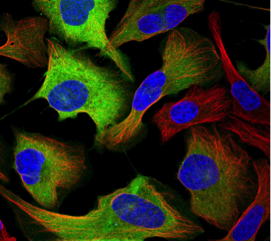
CCNB1: Cyclin B1 is a regulatory protein localized to the cytosol involved in mitosis in human and mouse cells. It is expressed in a cell cycle-dependent predominantly during G2/M phase of the cell cycle. In the image, cyclin B1 protein expression (antibody HPA030741), in the U-2 OS human cell line, is shown in green, while the nucleus is blue and microtubules in red.
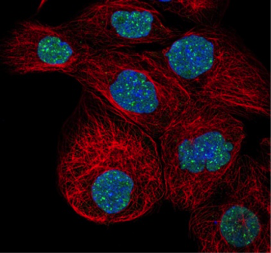
DAXX: Death-domain associated protein 6 is a multifunctional protein that is primarily localized to the nucleus and nuclear bodies in both human and mouse cells but also shows expression in the cytoplasm. DAXX associates with centromeres in G2 phase of mitosis. In the cytoplasm, it functions to regulate apoptosis through interaction with apoptosis antigen Fas. The image shows expression in the nuclear bodies within the nucleus (GFP-labeled antibody HPA008797) in cell line A-431.
(Images obtained from www.proteinatlas.org, accessed March 5, 2017; version 16.1)
Perhaps the most unique aspect of the new Cell Atlas is how the subcellular localization of each protein was identified. Online gamers were recruited to speed up the annotation through participation in Project Discovery. The Cell Atlas team, in collaboration with CCP Games, Massively Multiplayer Online Science (MMOS) and Reykjavik University created this mini-game within the popular EVE Online (a massively multiplayer online game) to “gamify” their research (www.eveonline.com/discovery/ ). The objective of game is for the EVE Online gamer to identify what cellular structure(s) display a fluorescent green color in the image they are presented. These can be roughly classified as within the nucleus, in the cell’s cytoplasm, or even outside the cell in the periphery. After completing a brief tutorial with pre-classified images, gamers are shown images to annotate in exchange for a small reward in the form of credits that can be used to purchase Sisters of EVE items. The results submitted by each player are compared with the results of other players to improve and expand a real-life database of information of human protein data. “At any time and place in EVE, players are able to play the mini-game “Project Discovery” and categorize the protein expression patterns from Cell Atlas images into different organelle categories, This was a help for us in classifying organelle substructures and refining the details in the Cell Atlas”, says Dr. Lundberg, director of the Cell Atlas.
The researchers expect the Cell Atlas to play a key role in the exciting new area of “spatial proteomics”. It is the systematic large-scale analysis of a cell’s proteins and their localization to distinct subcellular compartments, which is vital for deciphering a protein’s function(s) and possible interaction partners. Knowledge of where a protein spatially resides within the cell is important, as it not only provides the physiological context for their function but also plays an important role in furthering our understanding of a protein’s complex molecular interactions (i.e. signalling and transport mechanisms) by matching certain molecular functions to specific organelles (Breckels, 2016). This emerging field is predicted to dramatically broaden our understanding of human health and disease and having tools like the Cell Atlas will prove vital to this endeavor.
Footnotes
-
1. Breckels LM et al. Learning from Heterogeneous Data Sources: An Application in Spatial Proteomics (2016). PLoS Comput Biol 12(5): e1004920. doi:10.1371/journal.pcbi.1004920
-
2. Drain B (March 20, 2016). EVE Evolved: Why you should participate in EVE’s Project Discovery. Massively Overpowered. Retrieved March 5, 2017 from http://massivelyop.com/2016/03/20/eve-evolved-why-you-should-participate-in-eves-project-discovery/
-
3. Marx V (May 28, 2014). Proteomics: An atlas of expression. Nature 509: 645–649. doi:10.1038/509645a
-
4. KTH, Royal Institute of Technology. (January 22, 2015). First major analysis of Human Protein Atlas is published. ScienceDaily. Retrieved March 8, 2017 from www.sciencedaily.com/releases/2015/01/150122145415.htm
-
5. The Human Protein Atlas. About the Human Protein Atlas. Retrieved March 5, 2017 from http://www.proteinatlas.org/
-
6. The Human Protein Atlas. The Cell Atlas. Retrieved March 5, 2017 from http://www.proteinatlas.org/cell
-
7. Uhlén M et al. Tissue-based map of the human proteome (2015). Science 347 (6220). doi: 10.1126/science.1260419
-
8. Uhlén M et al. Towards a knowledge-based Human Protein Atlas (2010). Nature Biotechnology 28 (12): 1248–50. doi:10.1038/nbt1210-1248.