
A High Throughput, Scalable Solution to Upstream Process Development
A Guest Blog by Dr. Simon Dewar, Head of Upstream R&D Operations, FUJIFILM Diosynth Biotechnologies and Dr. Fern Slingsby, Lead Scientist, Upstream Development, FUJIFILM Diosynth Biotechnologies
Introduction
There are numerous challenges to be overcome when developing upstream manufacturing processes for the production of biotherapeutic drug products. High levels of process understanding and robustness are required for cGMP manufacture; however, the time taken to obtain these must be kept to a minimum in order to reduce costs and meet constraints such as asset slots and clinical requirements. New high-throughput upstream development approaches, therefore, are particularly desirable. However, it is also desirable that these approaches are scalable and so applicable to the manufacturing equipment and facilities for which the processes are being developed. This provides confidence that the results obtained – and so the decisions made – are relevant, otherwise the potential implications could be costly in terms of money, time and resources.
Current Approach to Upstream Development
Typically, upstream process development consists of strain generation and strain screening, followed by process optimisation. For microbial processes, strain screening is usually carried out in shake flasks and selections made based upon those strains showing the best productivity and product quality. However, shake flasks are often a poor approximation for bioreactors and allow for the screening of only a few parameters at best (e.g. temperature and inducer concentration). Parameters such as oxygen transfer are poor compared with larger vessels and conditions such as pH are neither closely monitored nor tightly controlled. Therefore, selection is unlikely to favour the best strains and conditions appropriate for larger fermenter systems.
Once the strains have been selected, development and optimisation of the fermentation process is carried out in bioreactors which are more representative of the vessels and conditions to be used during manufacturing. A wider range of controlled parameters can be investigated, such as media composition, feed profiles, pH, temperature, induction conditions and dissolved oxygen, to name but a few. This ensures that the microorganisms are responding to conditions representative of those to be used during manufacture, and as such the responses can be measured and decisions made based upon relevant fermentation conditions. However, running larger vessels is expensive, time consuming and requires a lot of resources in order to obtain enough process information to make informed decisions.
Using a statistically relevant Design of Experiments (DoE) approach can assist in ensuring that the effects of changing each parameter are detected in the best possible manner and yet using the minimum number of fermentations. However, this can still require a large number of fermentations and so is not only time consuming and expensive in the larger vessels, but often needs additional fermentations beyond the minimum required for the number of parameters to be investigated. This is due to “blocking”, which is often necessary due to the practicalities of running multiple, parallel fermentations and takes into account any potential variation caused by carrying out fermentations on different days, in different fermenters and/or with different operators. There is, therefore, a clear need for a scalable, reproducible, high-throughput solution to the challenges posed by strain screening and fermentation process optimisation discussed above.
The ambr250 ™ (+24) Parallel Bioreactor System

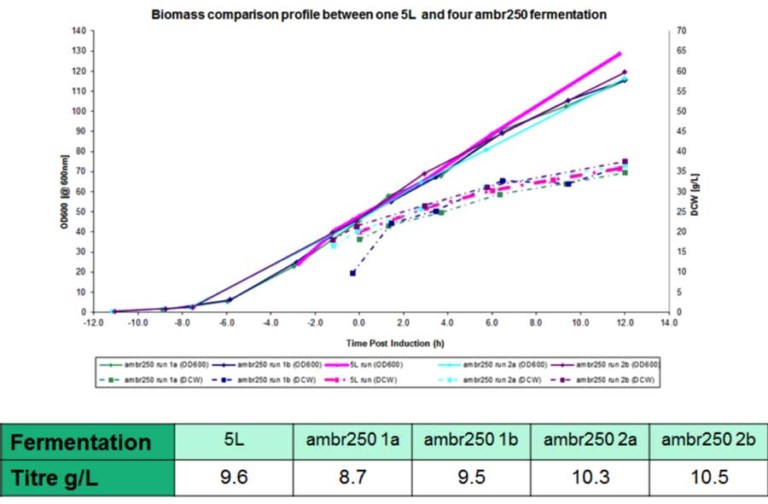
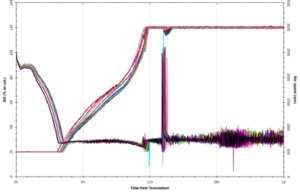
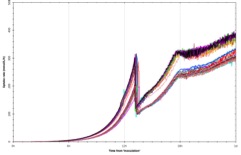
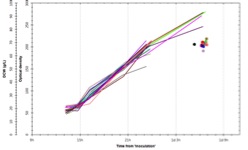
The system uses disposable scale-down versions of conventional bioreactors (complete with Rushton impellers, sparger, pH and DO probes, oxygen supplementation and up to four feed lines) and can run processes shown by FUJIFILM Diosynth Biotechnologies to scale up to at least 3000L – providing relevant process understanding and confidence for subsequent process scale-up and manufacturing. Evidence for this is given in Figure 2, showing growth profiles and product titres obtained when running a platform FUJIFILM Diosynth Biotechnologies’ Escherichia coli fermentation process using both the amb250 ™ system and conventional bioreactors, with comparable results obtained for each. Confidence in reproducibility is also demonstrated in Figure 3, where 23 vessels were run using an identical Fujifilm platform E. coli fermentation process. Again, comparable results were obtained for each. The ambr250 ™ system has continuous control and/or monitoring of parameters including pH, temperature, dissolve oxygen, off-gas analysis – in fact all the control and monitoring available for larger vessels and more. All sampling and additions can be automated, meaning the required operator time is kept to a minimum and disposable, pre-sterilised bioreactors virtual eliminate any risk of contamination.
Using the ambr250™ For Upstream Development
Using the ambr250 ™ system, strain screening can be carried out directly into fermenter vessels, thus eliminating the need for shake flasks. This enables the best strains to be selected for growth and expression in fermenter vessels, whilst allowing fermentation process optimisation to be carried out in parallel to strain screening, so high quality data and process understanding can be obtained much earlier on in the development process. In fact, using its proprietary pAVEway™ expression system, FUJIFILM Diosynth Biotechnologies is able to go from gene to defined fermentation process in a little as four weeks. Multiple DoE fermentations can be carried out simultaneously in a scalable, reproducible system. This ensures the maximum amount of statistically relevant data is collected using the minimum amount of runs (and also allows for additional runs to be carried out up to 24 per cycle – enabling greater process understanding). FUJIFILM Diosynth Biotechnologies has successfully used this system for combining a DoE approach with their pAVEway™ expression system and platform fermentation processes to obtain optimised, strain specific fermentation processes. The pAVEway™ system achieves tight control and modulation of expression, so in combination with platform fermentation processes and the ambr250 ™, rapid progression can be achieved from gene to high titre, strain specific, optimised fermentation processes which are robust, well characterised and suitable for cGMP manufacture.
Of course there is always the potential for high throughput systems such as the ambr250™ to create bottlenecks downstream, for instance in sample processing and data analysis, simply due to the amount of data and samples generated over a much shorter time frame when compared with typical development programmes. Conventional SDS-PAGE, for example, would not be practical for timely analysis of product expression in, say, four samples per 24 fermentations (or 96 per ambr250 run!). High throughput analysis tools such as the Octet and Caliper electrophoresis systems can help, enabling multiple samples to be processed in a fraction of the time taken for conventional analysis techniques. In addition, the vast amount of high-quality data created by the systems needs to be processed in order to ensure that the most benefit is obtained from the impressive capabilities of the systems. After all, what’s the point in collecting it if you’re not going to use it? Utilising the power of software packages such as JMP™ and Design Expert™ can help process the information and provide statistical analysis and visual representation of the large quantities of information obtained. FUJIFILM Diosynth Biotechnologies have developed bespoke scripts for effective mining of this information using JMP™and can apply multivariate analysis to further define the design space at an earlier stage in the development timeline. Obtaining such information early on in the development process not only facilitates a more refined fermentation process, but a more in depth understanding of manufacturing processes is advantageous from a regulatory point of view too, helping to prevent any unwanted surprises further on down the development pipeline.
Figure 3: Confidence in reproducibility is demonstrated below, where 23 vessels were run using an identical E. coli fermentation process. A – overlay traces of on-line dissolved oxygen (DO) and agitation (stir speed) for each vessel. B – overlay traces of oxygen uptake rate and carbon dioxide evolution rate for each vessel. C – overlay traces of off-line biomass measurements via optical density (OD600 nm) and dry cell weight (DCW) for each vessel.
Conclusions
In short, there is a clear need for reproducible, scalable high throughput approaches to upstream development during biotherapeutic production. This is to ensure relevant, high quality data is collected in a manner which reduces waste and ensures confidence when progressing processes for cGMP manufacture. The ambr250™ system meets these needs, with the ability to run 24 parallel fermentations with automated control and continuous monitoring in a system shown by FUJIFILM Diosynth Biotechnologies to run processes scalable up to at least 3000L. Combining this with high throughput analysis tools, design and analysis software packages (such as JMP™ and Design Expert™) and platform expression systems and fermentation processes (such as the FUJIFILM Diosynth Biotechnologies’ proprietary pAVEway™ expression system) ensures rapid progression from gene to optimised, high titre fermentation process with the in-depth process understanding and robustness required for cGMP manufacture of biotherapeutic drug products.
Author Bios:
Dr. Simon Dewar, Head of Upstream R&D Operations, leads a group of about 40 upstream scientists developing microbial and mammalian process for GMP manufacture. More than 25 years working in biotechnology business with a PhD from University of Manchester studying morphological evolution of filamentous fungi grown in continuous culture (Quorn).
Dr Fern Slingsby is a Lead Scientist within Upstream Development for FUJIFILM Diosynth Biotechnologies. She has over 10 years’ experience in protein production using a variety of expression systems, including molecular biology, fermentation and GMP manufacture. Dr. Slingsby has a PhD in Molecular Biology from the University of Edinburgh