Enzymes for Cellular Lysis or Protoplast Formation: Bacteria, Yeasts, and Plant
A guest post by Shanghao Li, Ph.D. is Global Product Manager of Immunology and Cell Biology at MP Biomedicals LLC
Digesting cell walls using enzymes is a critical component of many scientific approaches such as cellular lysis, proto/spheroplast formation, and numerous other approaches. The use of enzymes has advantages over physical or chemical methods because of its specificity, the use of milder conditions and lower shear stress.
Since cell walls differ in composition between types of cells, it is important to use an enzyme with the correct specificity and activity for the application. Other consideration in choosing an enzyme include the purity and need for other reagents or additional procedures related to the use of that particular enzyme.
Bacteria
Bacteria are categorized as Gram-positive or Gram-negative based on the cell wall structure. Peptidoglycan, which consists of glycan-tetrapeptide repeats connected by penta-glycine bridges, provides strength and rigidity to the cell wall.
Peptidoglycan has similar composition in both types of bacteria, however, in Gram-positive bacteria it is much thicker. Toward the outside of the Gram-positive cell, the peptidoglycan is connected to teichoic acids and polysaccharides [1].The wall of Gram-negative bacteria consists of an outer lipid membrane that covers peptidoglycan in a two-layer structure with the periplasmic space in between. The outer membrane of Gram-negative bacterial typically prevents access of digestive enzymes to the peptidoglycan. For that reason, Gram-negative cells typically require pre-treatment with a detergent or a chelating agent to destabilize or remove the outer membrane prior to digestion [1, 2].
Enzymes that cleave peptidoglycan are called murein hydrolases and are classified into 3 groups: glycosidases (cleaves polysaccharide chains), endopeptidases (cleaves polypeptide chains), or amidases (cleaves between polysaccharides and peptides) [1, 2].
Lysozyme
Among muramidases, lysozyme is the most widely used as it has activity against most Gram-negative and many Gram-positive bacteria. Hen egg white lysozyme is the most commonly used lysozyme because of its activities, availability, and cost. Extensive hydrolysis of peptidoglycan by lysozyme may be sufficient for cellular lysis without additional measures in some bacteria. In others, an additional procedure, such as a hypotonic shock and prior sensitization, may be required for complete lysis. Lysozyme treatment of viable cells of some bacteria can prove more difficult.
Lysozyme is commonly used for the lysis of E. coli, Streptomycetes, and other bacteria. In addition, there are numerous other uses beyond bacteriolysis and protoplast formation such as antimicrobial control, sample preparation, pharmacology, as well as a flavor enhancer in some foods and drinks.
Other Bacteriolytic Enzymes:
Other enzymes are available for bacteria that are resistant to lysozyme (such as many Gram-positive bacteria) or in cases where an alternative to lysozyme is desired. Examples include:
- Lysostaphin is an endopeptidase with specificity for the pentaglycine bridges of peptidoglycan. It has been used with Staphylococci strains and as a potential antimicrobial [4].
- Achromopeptidase is a broad-spectrum endopeptidase with specificity for Lys/- bonds that retains activity in 0.1% SDS and 5M urea. It has been used for the preparation of Actinomycetes protoplasts and for lysis of Staphylococcus and Micrococcus [5].
- Labiase has β-N-acetyl-D-glucosamidase and muramidase activities and been shown to have broad-spectrum bacteriolytic activity on gram positive bacteria [6]. It has been used for DNA extraction and other uses.
- Mutanolysin was first characterized as composite of 3 enzymes from S. globisporus with β-1,4-N,6-O-diacetylmuramidase (M1), β-1,4-N-acetylmuramidase (M2), and N-acetylymuramyl-alanine amidase activities. The isolated M1 enzyme may also be referred to as Mutanolysin [7]. It has been used for the lysis of Listeria, Lactococcus, Lactobacillus, and Streptococcus, and has been used for protoplast formation and DNA isolation.
Yeast
Digesting yeast cells walls is needed in applications such as include nucleic acid extraction, transformation, protoplast formation, cell fusion and numerous others. The use of yeast-lysing systems is often an early step for recovering recombinant protein expressed in yeast.
The use of lytic enzymes for cell disruption in protein recovery avoids some of critical drawbacks that mechanical methods have such as high viscosity, proteolysis, contamination, and shearing. Through the use of purified, protease-free enzymes it has been possible to carry out controlled Saccharomyces cerevisiae lysis, to result in the selective release of cloned (recombinant) intracellular protein particles [8]. The specific activity of the lytic enzymes is a key factor to be considered as it must sufficiently high to obtain fast cell breakage without allowing endogenous intracellular proteases to degrade the product.
Yeast Cell Walls
The outer surface of yeast cell walls is composed mostly of mannoproteins, which are densely packed and limit permeability to the glucans of the inner wall. The inner wall glucans consist primarily of fibrous β-1,3 glucan scaffolds with some β-1,6 glucans which links the components of the inner and outer walls. The glucans provide strength, shape, and elasticity to the wall [1].
These components are covalently linked into macromolecular complexes, which are assembled in unit modules built around a molecule of β-1,3 glucan and sometimes β-1,4 chitin [1]. The modules are further linked by noncovalent interactions in the glucan–chitin layer and by cross-linkages and disulfide bonds between mannoproteins [1].
Yeast-Lysing Enzyme Systems
Enzyme lysis systems for yeast cellular lysis typically consist of one or more enzymes (which act synergistically) which may include protease, β-1,3 glucanase (lytic and nonlytic), β-1,6 glucanase, mannanase, and chitinase [1].
The protease functions to open the protein structure and expose the inner glucan surface to attack from a glucanase, which then solubilizes the glucan. In some cases, reducing compounds can disrupt mannoprotein and effectively replace the protease [9, 10]. Once the glucanase(s) has opened a sufficiently large hole in the cell wall, the plasma membrane and its content are extruded as a protoplast. In osmotic buffers of sucrose or mannitol, the protoplast remains intact, but in hypotonic solution it lyses immediately.
Zymolyase is an enzyme preparation from a submerged culture of the soil bacterium Arthrobacter luteus (also known as
Cellulomonas cellulans) which effectively lyses cell walls of viable yeast cells [11]. Zymolyase contains a group of enzymes. The primary enzyme is ß-1,3-glucan laminaripentaohydrolase which hydrolyzes glucose polymers at the ß-1,3-glucan linkages to release laminaripentaose as the primary product [11];. Zymolyase also includes protease and mannanase to strongly degrade yeast cell walls [11].
The lytic spectrum of Zymolyase include organisms from the following genera: Ashbya; Candida; Debaryomyces; Eremothecium; Endomyces; Hansenula; Hanseniaspora; Kloekera; Kluyveromyces; Lipomyces; Metschikowia; Pichia; Pullularia; Torulopsis; Saccharomyces; Saccharomcopsis; Saccharomycodes; Schwannimomyces and others.
Lyticase is a preparation from the same/similar organism [10]. Users report that Lyticase may be beneficial in many applications because of its different activity profile; however, many users report lyticase is less active for the complete cellular lysis of Saccharomyces. As demonstrated in experiments, zymolyase can easily break down various yeast cell wall components at significantly higher efficiency but lower concentration of the enzyme, enabling maximal yield of viable protoplasts (see Figure 1).
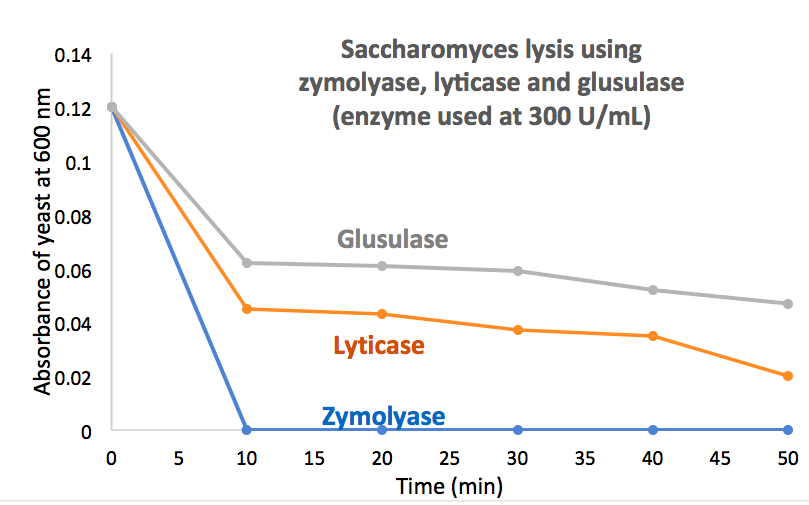
In applications where cellular lysis must be performed quickly and completely, such as isolation of an expressed protein, the use of a high specific activity preparation can be highly beneficial. MP Biologicals markets a lyophilized Zymolyase partially purified by affinity chromatography that contains 100,000 units enzyme/g.
Plants
Protoplasts of plants are widely needed for DNA transformation, plant breeding, and other uses. Protoplasts can be produced from a variety of plant species, plant structures, or plant cultures. Protoplasts can fully regenerate into functional plants when given proper plant growth regulators and growth medium.
Plant protoplast can be generated by using either one-step or two-step procedures. In the two-step method, cells are first macerated (separated) with pectinase(s) (or pectolyase) prior to conversion into protoplasts by cellulase treatment [12]. In contrast, the one-step process uses a complete enzyme cocktail to macerate and degrade the walls [13]. The one-step process, while convenient, requires detailed balancing of the two enzyme functions and prior optimization of the process.
The enzymes used for the isolation of protoplasts are typically extracts of fungal origin. The pectinases are rich in polygalacturonidase activity while commercial cellulase preparations may have multiple beneficial activities that include β -1,4-glucanase, hemicellulase, chitinase, lipase, nuclease, xylanase, and others [14].
Pectinases
During maceration, the breakdown of pectins (pectin acids and hydroxyls) leads to a loss of cohesion and cell separations. Both Endo-polygalucturonase or endo-pectate lyases have been reported to macerate specific tissues [15]. However, mixtures of the two activities have found a preference in the scientific literature and most commercial preparations contain a mixture of the activities.
Pectinase preparations can be derived from numerous genera, including Bacillus, Aspergillus, and Rhizopus. MP Biomedical supplies pectinase from Aspergillus niger with a specific activity of >1000 u/g.
Pectolyase is specific preparation from Aspergillus japonicas [15]. It has been characterized to contain endo-polygalacturonase(s) and endo-pectin lyase(s) in high activity in addition to a maceration simulating factor[16]. Pectolyase Y-23 is a specific high-activity commercial preparation form A. japonicus that has found wide use and acceptance in the scientific literature [17]. MP Biologicals supplies both of these with an activity of > 1000 u/g.
Cellulases
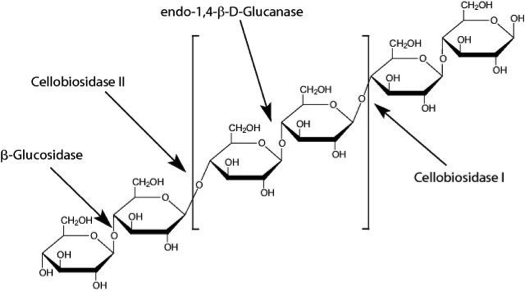
Similar to pectinases, cellulases comprise any of broad array of enzymes that hydrolyze the 1,4-beta-D-glycosidic linkages in cellulose, hemicellulose, lichenin, and other substrates. Cellulase can be produced by fungi, bacteria or protozoans.
There are 5 main classes of cellulases:
- Endocellulases randomly cleave internal bonds.
- Exocellulases produce di- or tetrasaccharides from the ends of the chains.
- Cellobiases or β-glucosidases further hydrolyse the exocellulase product to form individual monosaccharides.
- Oxidative cellulases depolymerize cellulose by radical reactions.
- Cellulose phosphorylases depolymerize cellulose using phosphates rather than water.
Cellulase Y-C from MP Biomedicals is produced form Trichoderma viride and has very high filter paper decomposing activity as well as appreciable additional xylanase and hemicellulase activity. It is an effective enzyme for use with Pectolyase Y-23 for plant cell wall removal. It has been used in numerous studies including the regulation of the cellulase transcription factors the liquefaction of hydrothermally pretreated wheat straw [18].
Cellulases also have numerous other uses such as food processing, agriculture, feed production, textiles, paper and biofuel production.
For more information, please visit https://www.mpbio.com/
References
- Salazar, O. and J.A. Asenjo, Enzymatic lysis of microbial cells. Biotechnol Lett, 2007. 29(7): p. 985-94.
- Harrison, S., Bacterial cell disruption: a key unit operation in the recovery of intracellular products. Biotech. Adv., 1991. 9: p. 217-40.
- Andrews, B.A. and J.A. Asenjo, Enzymatic lysis and disruption of microbial cells. TIBTECH, 1987. 5: p. 273-77.
- Recsei, P.A., A.D. Gruss, and R.P. Novick, Cloning, sequence, and expression of the lysostaphin gene from Staphylococcus simulans. Proc Natl Acad Sci U S A, 1987. 84(5): p. 1127-31.
- Li, S., S. Norioka, and F. Sakiyama, Bacteriolytic activity and specificity of Achromobacter beta-lytic protease. J Biochem, 1998. 124(2): p. 332-9.
- Niwa, T., et al., Lytic enzyme, labiase for a broad range of Gram-positive bacteria and its application to analze functional DNA/RNA. Microbiol Methods, 2005. 61: p. 251-260.
- Bronneke, V. and F. Fiedler, Production of bacteriolytic enzymes by Streptomyces globisporus regulated by exogenous bacterial cell walls. Appl. Environ, 1994. 60(3): p. 785-91.
- Asenjo, J.A., et al., Selective release of recombinant protein particles (VLPs) from yeast using a pure lytic glucanase enzyme. Biotechnology (N Y), 1993. 11(2): p. 214-7.
- Ventom, A.M. and J.A. Asenjo, Two extracellular proteases from Oerskovia xanthineolytica LL-G109. 1990.
- Scott, J.H. and R. Schekman, Lyticase: endoglucanase and protease activities that act together in yeast cell lysis. J Bacteriol, 1980. 142(2): p. 414-23.
- Ferrer, P., Revisiting the Cellulosimicrobium cellulans yeast-lytic beta-1,3-glucanases toolbox: a review. Microb Cell Fact, 2006. 5: p. 10.
- Nagata, T. and I. Takebe, Cell wall regeneration and cell division in isolated tobacco mesophyll protoplasts. Planta, 1970. 92(4): p. 301-8.
- Power, J.B., S.E. Cummins, and E.C. Cocking, Fusion of isolated plant protoplasts. Nature, 1970. 225(5237): p. 1016-8.
- Smith, H., The manipulation of plant cells, in The molecular biology of plant cells, H. Smith, Editor. 1978, Univ. of California Press.
- Ishii, S., Enzymatic maceration of plant tissues by endo-pectin lyase and endo-polygalacturonase from Aspergillus japonicus. Phytopathology, 1976. 66: p. 281-289.
- Baldwin, E.A. and R. Pressey, Pectic enzymes in pectolyase: separation, characterization, and induction of ethylene in fruits. Plant Physiol, 1989. 90(1): p. 191-6.
- Winters, A., S. Smeekens, and A. Cairns, Fructosyl transferase activity in the tissue-macerating preparation, pectolyase Y-23: Phsiological role of furctosyl transfer in Aspergillus and significance for studies of fructan synthesis in grasses. New Phytol., 1992. 121: p. 525-533.
- Silva, C. and E. Filho, A Review of holocellulase production using pretreated lignocellulosic substrates. Bioenergy Res, 2017. 10(2): p. 592-602.