
Implementation of the Sartorius Octet® BLI Platform Accelerates Early Antibody Discovery Workflows
Therapeutic monoclonal antibodies (mAbs) are a predominant class of new drugs directed against an array of target antigens used to treat cancers and immunologic diseases. The discovery process for antibody-based drug candidates includes several steps from target ID and validation, hit identification to preclinical lead candidate selection prior to clinical trial testing to evaluate efficacy and safety. On average, development time for an antibody drug candidate can take 12–15 years and cost more than $1 billion. Solutions are needed to increase the efficiency and throughput of the screening, characterization and target identification in early antibody discovery to drive down costs and development time.
In a webinar entitled Integrating the Octet® into Early Antibody Discovery Workflows, Dr. Colby Souders, Chief Scientific Officer at Abveris discusses how integrating the Octet® Bio-Layer Interferometry (BLI) platform (Sartorius) can help accelerate early antibody discovery workflows by enabling rapid selection and in-depth characterization of lead candidates. The Octet® BLI platform is a label-free biosensor-based solution capable of analyzing a variety of biological binding interactions, from mAb titer determination, to screening binding affinity of mAbs to their target receptors, and assay development to rapidly assess the quality of the new modality antibodies to improve drug discovery outcomes. Here, we will give a brief overview of the key messages and summarize several use cases as well as in vivo studies detailed in the webinar.
Dr. Souders began by outlining the current challenges in the drug discovery process for biologics where extremely low success rates (on average 11.5%), with failures skewed towards late-stage clinical phase testing. There is an identified need for function forward discovery approaches during candidate down selection to minimize the number of campaigns required to identify leads and increase the likelihood of preclinical candidates succeeding in the clinic. Dr. Souders emphasized that both high throughput and high resolution screening platforms are needed. At Abveris, advanced Octet® screening is used throughout two main upstream antibody discovery platforms including:
- Hybridoma discovery process (Abveris DiversimAb™ mice)
- Single B cell discovery process
While they still rely on traditional ELISA screening methods, Abveris also implement higher resolution methods such as the iQue flow cytometry system. But the main advantage of the Octet® platform is it combines both high throughput and resolution screening capabilities. The ability of the Octet® to perform analysis on crude supernatants and non-purified samples provides advantages to reduce processing time enabling rapid and reliable execution of quantitation, kinetics, and binning assays to characterize lead candidates or hits in downstream workflows.
In one example, the real-time, higher resolution advantage of the Octet® is apparent when compared to traditional ELISA assays in both an indirect format and the more predictive capture format matched to the Octet® setup. Based solely on ELISA data alone, there is very little discrimination the antibody clones making it difficult to rank them effectively. The Octet® can provide information on binding affinities and kinetics profiles, which the ELISA does not, as well as the resolution needed to discriminate between the clones. When the off-rate ranking is compared with ELISA results, there is distinct clustering of three clones (11E2, 9G10, and 3D9) that have very different kinetic profiles despite their similar performance in ELISA.
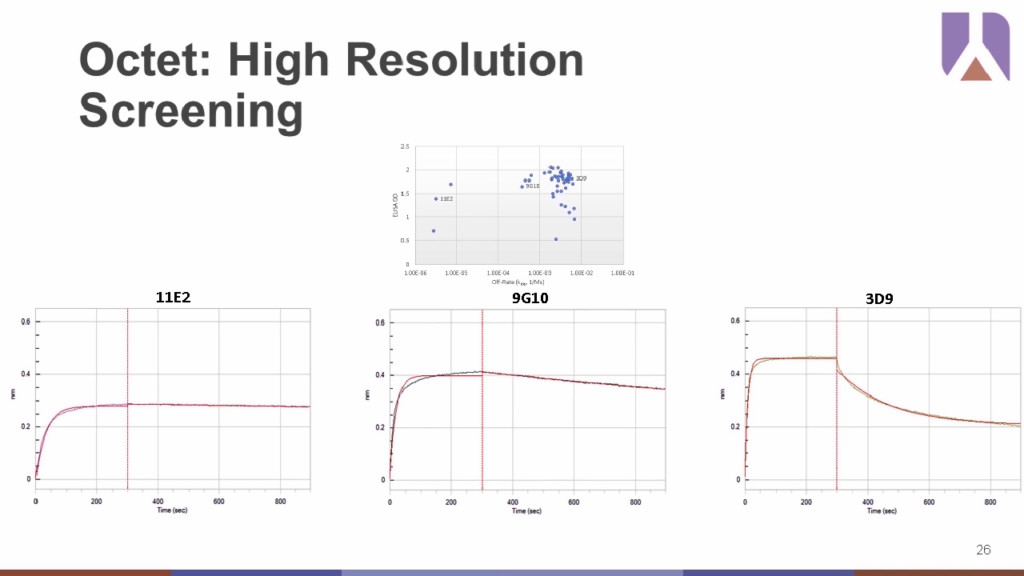
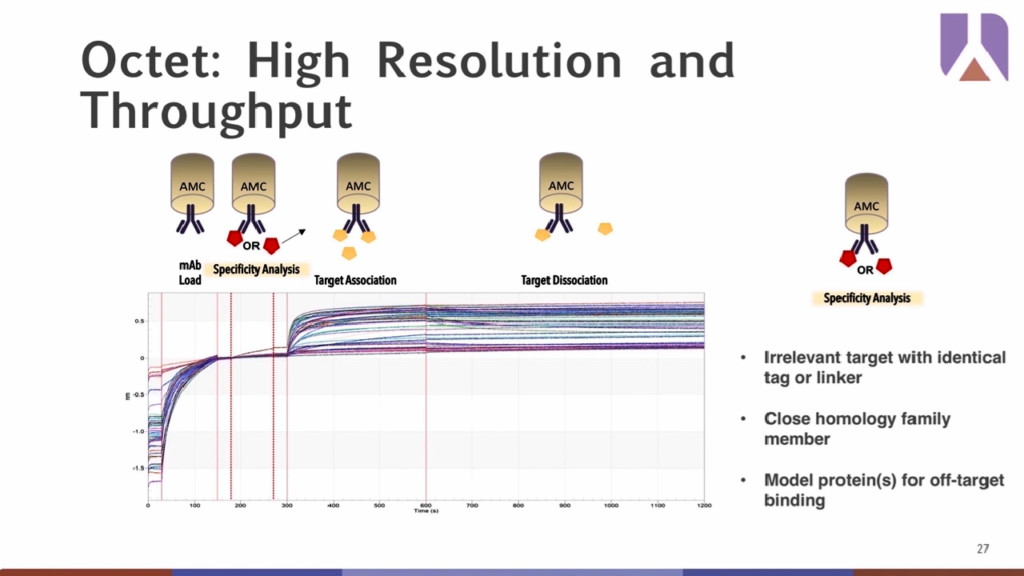
Moreover, the sensitivity of the Octet® enables easier detection of unwanted interactions, such as irrelevant targets with identical tags or linkers, close homology family member, or allow for the modeling of proteins that can predict off-target developability problems downstream.
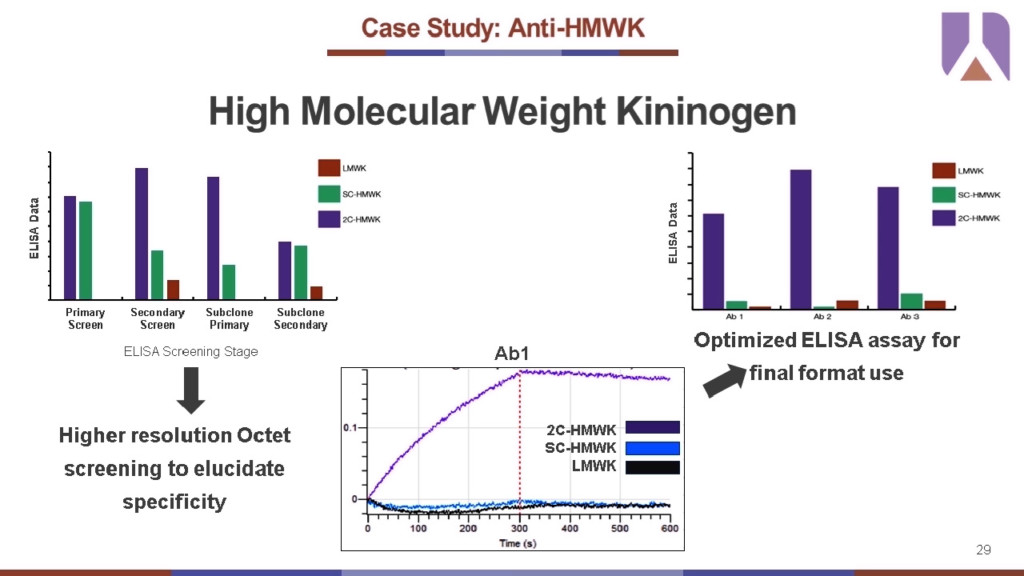
CASE STUDY 1: Anti-High Molecular Weight Kininogen (Anti-HMWK)
The sensitivity of the Octet® screening was critical to success of an anti-HMWK discovery campaign specifically to the cleaved isoform. The kininogen protein itself is a circulating plasma protein involved in contact pathway activation and coagulation. It is cleaved by prekallikrein to release the small peptide bradykinin leaving behind a two-chain (2C) cleaved version of HMWK that is of interest as a promising biomarker target for antibody discovery. This is a challenging target because it is a small, conformationally constrained epitope due to the requirement of a post-translational cleavage process and the presence of multiple cleaved variants.
The standard ELISA screening results at various stages of discovery couldn’t identify mAb clones with high specificity, but high-resolution screening of a large antibody panel by Octet® successfully identified several clones with specificity for 2C-HMWK (over single chain) binding. The top three candidates identified by the Octet® screening helped narrow down optimization efforts and allowed Abveris to design an ELISA format that could characterize this specificity for final format use.
Beyond specificity alone, Dr. Souders emphasized the importance of functional characterization in therapeutic mAb discovery campaigns, such as an antibody’s ability to neutralize receptor-ligand interactions to modulate signaling cascades. The Octet® can be used to measure the mAb kinetic profiles against multiple targets and to assess the neutralizing capabilities of the antibody-receptor complex by dipping the Octet® probe into a solution containing the receptor’s ligand. Based on the target epitope of interest, this analysis can help find neutralizing or non-neutralizing mAb clones (binds to the receptor but still allows for ligand binding).
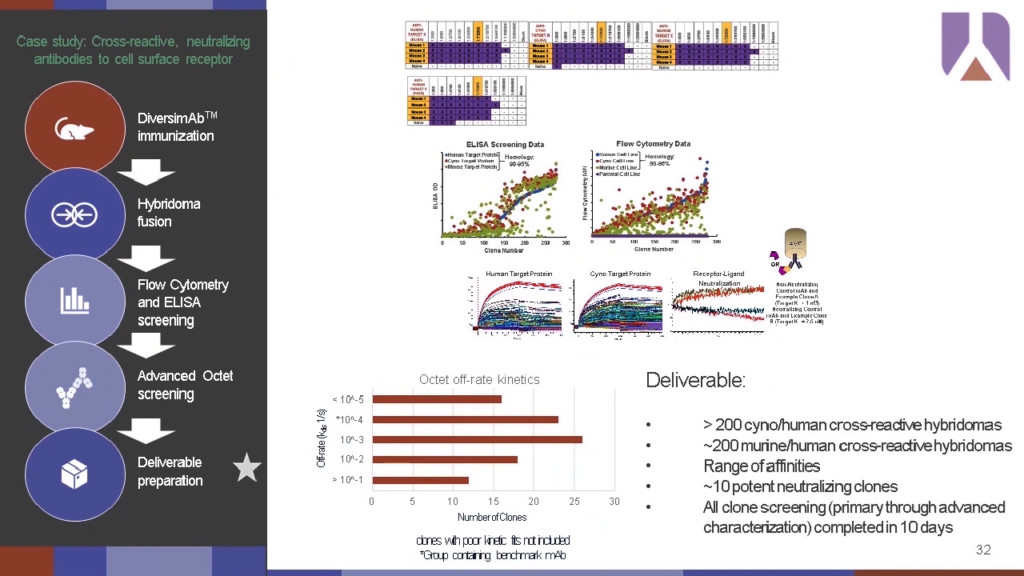
To better illustrate how this high-resolution screening is critical for lead candidate down selection, Dr. Souders presented several examples using the Abveris DiversimAb™ screening platform. In one instance, they were looking for a cross-reactive and neutralizing Ab to a cell surface receptor target. Cross-reactive clones were evaluated with advanced Octet® screening to determine kinetic profiles and find clones that demonstrated comparable non-neutralizing behavior in a receptor-ligand neutralization assay with similar performance to benchmark controls. This workflow was able to advance quickly with all the screening from primary data to advanced Octet® characterization completed in 10 days to down-select from 200 clones down to 10 lead candidates and allowed the team to focus downstream efforts solely on candidates with the desired properties.
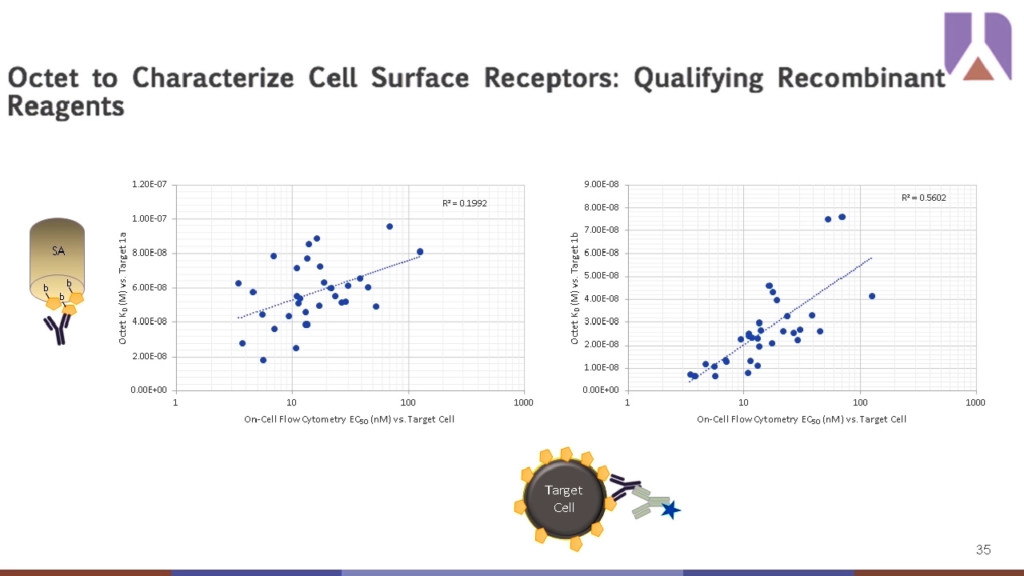
Another example of the Octet®’s abilities extend to assessing the quality of recombinant reagents in discovery workflows. Multi-pass transmembrane protein targets present challenges for antibody discovery because it can be difficult to recapitulate their quaternary structures in vitro for screening purposes. Recombinant analogs need to represent on-cell affinities to be used predictively in large-scale mAb discovery campaigns. The Octet® can be used to screen small panels of recombinant reagents to determine the best candidate to use for larger therapeutic campaigns and downstream characterization for final lead candidates. It is important to have good correlation between the native and recombinant versions of the target protein since downstream engineering efforts generally rely heavily on in vitro selection techniques. In the example below, two recombinant versions of the target protein where Octet® affinity data (Y-axis) for each compared to the on-cell apparent affinity (X-axis) as measured by flow cytometry. Of the two versions, 1b shows better correlation between the Octet® and on-cell data (R2=0.5602) and is the better recombinant protein to use in large-scale screening.
CASE STUDY 2: Engineering of Lead Candidates
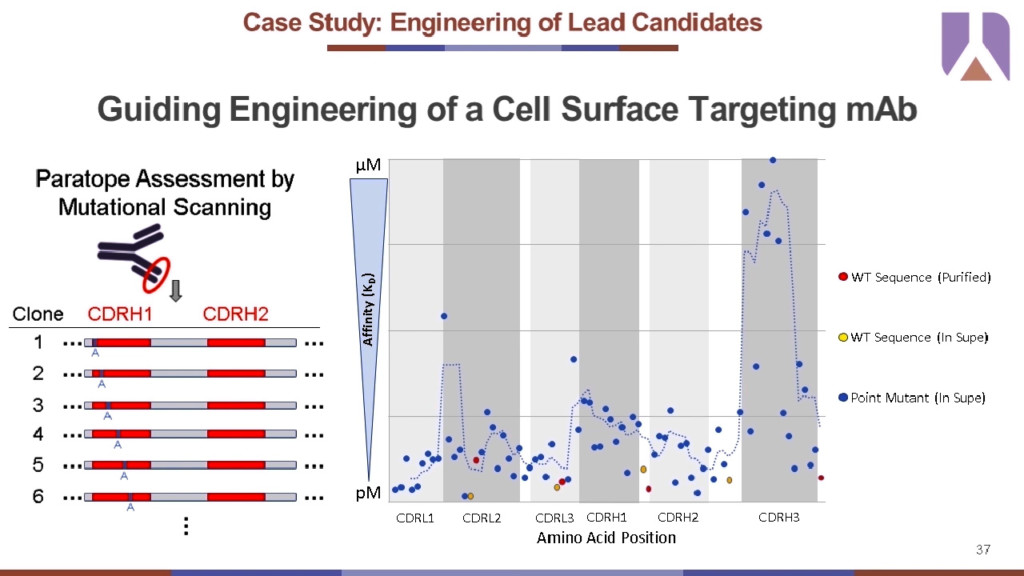
In this case study, Abveris had identified a high-quality recombinant protein and were interested in engineering for improved characteristics. Here, performing an initial paratope analysis can help narrow down the region of interest for mutagenesis library generation. Traditionally, a full mutagenesis library can only be generated at a limited number of amino acid positions per library due to diversity constraints and it is laborious to create these screening libraries. So, constructing and screening initially against a panel of single amino acid point mutations at all regions of interest can narrow down the residues to select for full library generation. For example, each amino acid position in all complementarity-determining regions (CDRs) of an antibody were iteratively mutated to alanine to generate a mini library of single point mutations. The antibody binding is dependent on a handful of critical residues in the heavy chain CDR3 (CDR3H) based on the affinity values (Y axis) compared against the position of the point mutation in the antibody sequence (X axis). This data can be used to mature antibody development in many ways depending on the goals. For instance, to improve developability without affecting critical affinity residues, you could narrow down the full mutagenesis library to target heavy chain CDRH1 and CDRH2.
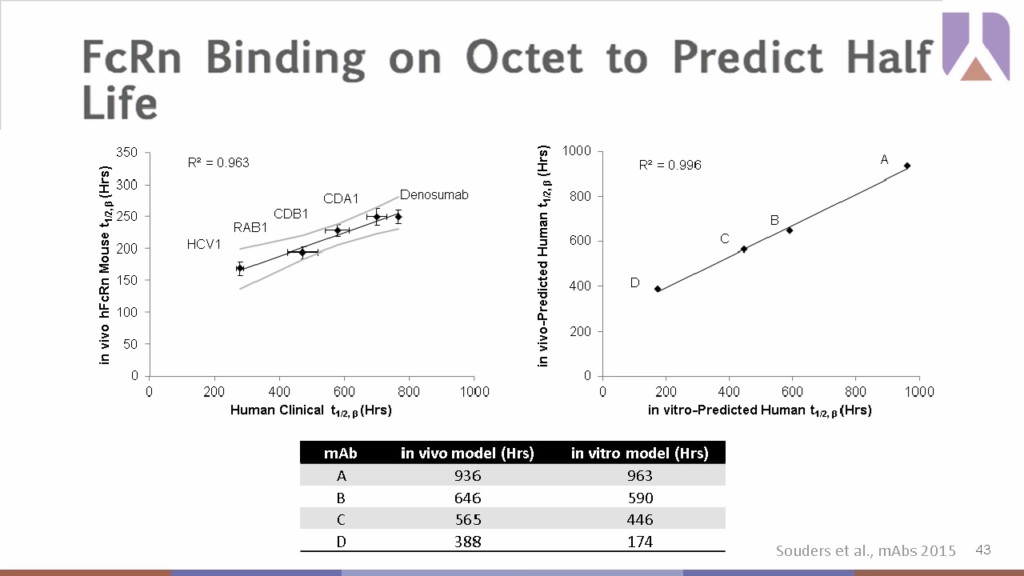
Once preclinical candidates have been narrowed down, evaluating critical binding interactions prior to in vivo studies can enhance downstream clinical success. The Fc region of human IgG contributes to the biological and pharmacological characteristics of therapeutic antibodies, most notably it impacts the antibody plasma half-life and in turn, overall drug efficacy. The pH dependent binding interactions of the Fc region to neonatal Fc-receptor (FcRn) can increase or decrease serum half-life of Fc-containing therapeutics, so effective modeling of Fc-FcRn binding interactions can allow for ranking of antibodies to select those with longer predicted half lives for downstream assessment. The Octet® predicted antibody half-life based on Fc-FcRn binding interaction was compared against results from in vivo mouse models to validate the in vitro model. Across four experimental mAbs, there was a strong correlation between the in vitro and the in vivo results providing confidence in the predictive power of the Octet® platform. Dr. Souders noted that this in vitro model can greatly expedite characterization and subsequent down selection based on half-life characteristics providing the added benefits of higher throughput and ease of execution over in vivo testing, where time-to-data is slow.
The case studies presented by Dr. Souders effectively illustrate how enabling technology solutions such as the Octet® BLI platform, as a replacement or supplemental tool, for a variety of antibody discovery protocols (kinetics, affinity analysis and binding specificity) can enable rapid down selection of potential lead candidates in discovery campaigns. The right combination of throughput and resolution can improve overall drug development success rate by allowing researchers to find and characterize the best candidates as early in the screening process as possible. Reducing the number of campaigns required by advancing high quality candidates allows for faster, more cost-effective movement through discovery workflows to drive better clinical outcomes. Moreover, as the therapeutic antibody field explores new modality antibodies such as bispecific and trispecific antibodies and antibody-drug conjugates, rapid and robust quantitation, and functional characterization, as offered by the Octet® BLI platform, will be critical in the speed to bring them to market.
For more information, please see:
To view the webinar in full, please visit: Integrating the Octet® into Early Antibody Discovery Workflows