Materials and Assay Systems Used for 3D Cell Culture
Guest Blog By: Suparna Sanyal, Ph.D., Scientific Writer and Marshall Kosovsky, Ph.D., Global Technical Support Manager, Corning Life Sciences
Introduction
Mammalian cell culture has served as an invaluable tool for cell biology research. Monolayers of adherent cells grown on flat and rigid two-dimensional (2D) substrates, such as polystyrene or glass, have evolved as a standard approach for conventional cell culture. 2D cell culture studies have played a pivotal role in furthering our understanding of developmental biology, tissue morphogenesis, disease mechanisms, drug discovery, large-scale protein production, tissue engineering, and regenerative medicine. However, there are inadequacies and limitations associated with 2D culture systems, especially regarding their inability to emulate in vivo conditions and provide physiological relevance.
In the body, nearly all cells in tissues reside in an extracellular matrix (ECM) consisting of a complex three-dimensional (3D) architecture, and interact with neighboring cells through biochemical and mechanical cues(1). Cell-cell and cell-ECM interactions establish a 3D communication network that maintains the specificity and homeostasis of the tissue(2). Key events in the life cycle of a cell are regulated by organizing principles that are determined by the surrounding cellular microenvironment(3). The inability of cells to achieve in vivo-like structural organization and connectivity in 2D cell culture assays can limit or diminish properties such as cellular morphology, viability, proliferation, differentiation, gene and protein expression, response to stimuli, drug metabolism, and general cell functionality.
To overcome some of these shortfalls, numerous 3D cell culture models have been developed over the past two decades to address the diverse requirements of different cell types and applications. Emerging evidence strongly suggests that 3D cell cultures that establish physiological cell-cell and cell-ECM interactions can mimic the specificity of native tissue with greater physiological relevance than conventional 2D cultures(8-10). According to a 3D cell culture trends report(11), natural ECM-based hydrogels (e.g., Corning® Matrigel® matrix and Collagen) and 3D cell aggregates known as spheroids are among the most widely used models for 3D culture in vitro. These 3D models have been used to demonstrate physiologically relevant properties of diverse cell types, and have been extensively applied stem cell differentiation, tumorigenesis, and drug discovery research. Spheroids and/or natural hydrogels are advantageous since they can be manipulated into more complex in vivo-like co-culture models that incorporate multiple cell types. Furthermore, devices such as permeable supports (e.g., Transwell® inserts) can be incorporated into 3D culture models with a hydrogel substrate(s) to facilitate the analysis of interactions between different cell types, soluble factors, and the culture microenvironment.
1. 2D versus 3D Cell Culture Models
Fundamental differences in the microenvironment of 2D and 3D cell culture systems influence various cellular behaviors including cells attachment, growth, morphology and polarity, gene and protein expression, response to stimuli, cellular metabolism, and overall function. One of the primary differences observed when comparing cells in 2D and 3D cultures is the dissimilarity in cell morphology. Cells adopt 2D or 3D confirmation primarily based on the orientation of integrin-mediated adhesions to the extracellular matrix. In the case of 2D cultures, cell attachment occurs on one side of the cell (the side that contacts the 2D surface), whereas 3D cell culture promotes attachment around the entire surface of the cell(13). In general, cell attachment and spreading on restraint-free 2D substrates occurs within minutes. In contrast, cell attachment and spreading in 3D culture systems is preceded by proteolytic degradation of their physical environment, which can occur over the course of hours, or even days in some instances(18).
It has been suggested that the degree of cell spreading can impact cell proliferation, apoptosis and differentiation(19-22). Many cells, when isolated from tissues and placed onto planar cell culture surfaces, become progressively flatter, divide aberrantly, and lose their differentiated phenotype(23,24). Interestingly, some of these cell types can regain their physiological form and function when embedded in a 3D culture environment. For instance, encapsulation of dedifferentiated chondrocytes in 3D cultures restores their physiological phenotype, including cell shape and the expression of cartilaginous markers(25). Similarly, mammary epithelial cells embedded in a 3D environment halt uncontrolled division, assemble into acinar-like structures, and establish a de novo basement membrane(24,26,27).
Another important physiological attribute conferred by 3D models is appropriate cell polarity. Polarity in vivo depends both on the cell type and the tissue microenvironment. Epithelial cells are often polarized, with apical and basolateral surfaces that are important for tissue organization and directional secretion of bioactive molecules. Tissue organization is lost when these cells are explanted onto flat 2D tissue culture substrates. When they are returned to appropriate 3D culture conditions, epithelial cells generally regain apical-basolateral polarity, and glandular cells form a lumen into which cellular factors are secreted(35). Permeable supports, such as Transwell Inserts, have been particularly useful for recreating 3D models of epithelial cells with native tissue-like cell morphology, cell-cell interaction, polarity, and secretory function(39-42).
Gene expression and mRNA splicing patterns can also vary considerably depending on whether cells are cultured under 2D versus 3D conditions(43,44). For example, melanoma cells exhibit distinct gene expression patterns when cultured on 2D substrates, in contrast to the properties observed with melanoma cell spheroids cultured in a 3D environment. Genes up-regulated in the 3D spheroids are also found to be up-regulated in tumors in vivo(45). When mammary epithelial cells are cultured on 2D substrates, these cells exhibit a dramatic up regulation of α1 integrin mRNA expression. In contrast, when these cells are cultured under 3D conditions using a reconstituted basement membrane such as Corning Matrigel matrix, the cells exhibit mRNA expression levels comparable with those observed in native breast tissue (reviewed in 13,46).
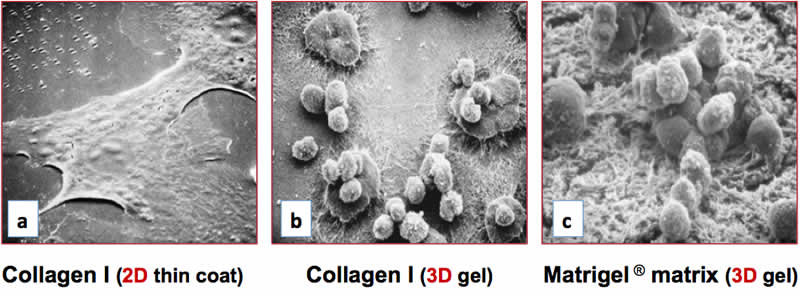
The differentiation niches of primary cells and stem cells are inherently 3D, and their biochemistry and topology have been found to dramatically affect the differentiation process(47). For example, primary hepatocytes cultured as a monolayer dedifferentiate and die within a few days(48). Dedifferentiated hepatocytes lose the ability to synthesize drug-metabolizing enzymes that are essential for toxicity assays in pharmaceutical research(48). The deficits in hepatocyte function observed on 2D surfaces can be overcome by embedding primary hepatic cells within 3D matrices such as Collagen I(49), Corning Matrigel matrix(49), synthetic peptide scaffolds(50), or by maintaining them under conditions of perfusion flow(51). Hepatocytes cultured on 3D matrices of collagen I or Matrigel Matrix form clusters and exhibit a rounded morphology that is indicative of the differentiated state (Figure 1b, 1c), yet were found to adopt a flattened, dedifferentiated morphology on a 2D collagen I substrate (Figure 1a). Under these conditions, hepatocytes cultured on 3D substrates exhibited optimal expression of the liver-enriched transcription factor C/EBP and elevated levels of cytochrome P450 enzymes, and were found to exhibit markedly reduced activities on the 2D surface (data not shown). Collectively, these observations demonstrate that 3D cell culture systems are capable of supporting varying degrees of cell complexity and functionality that are observed in vivo, which is dependent on the cell type and culture conditions. In contrast, 2D culture models usually support limited cell differentiation and in vivo-like functionality.
2. 3D Cell Culture Techniques
A critical need for in vitro 3D cell culture systems is to effectively mimic specific aspects of in vivo cell behavior to provide relevant models for studying tissue development and morphogenesis, cellular differentiation, genotypic, and/or phenotypic response to compounds in drug and toxicity screening assays. Some basic 3D models have involved the culture of cellular aggregates in suspension without the use of matrix-based substrata. However, the majority of more complex 3D cell culture models utilize either hydrogel-based matrices or solid scaffolds. A vast array of materials and fabrication techniques has been employed to develop scaffolds with varying physical and biological characteristics to address the requirements of different cell types. Of the current materials in use, naturally derived ECM-based hydrogels are most commonly used for in vitro applications of 3D cell culture.
3D Spheroid Cultures
Cellular spheroids are simple 3D models that can be generated from a wide range of cell types, which form spheroids because of the tendency of adherent cells to aggregate. Common examples of spheroids include embryoid bodies, mammospheres, tumor spheroids, hepatospheres, and neurospheres. Adherent cells have a natural tendency to aggregate and form spheroids under circumstances that impede adhesion to cell culture substrates. Common matrix-free methods employed for generating spheroids include the use of attachment resistant cell culture surfaces such as the Corning® Ultra-Low Attachment surface, or by maintaining the cells as suspension cultures in media (e.g., hanging drop technology, rotary cultures, and bioreactors). Several cell types also form spheroids in 3D hydrogels, and to a limited extent, in some solid scaffolds depending on the structural and physical properties of the material. The overall size of spheroids is limited to a few hundred micrometers, beyond which, necrosis ensues within the core of the spheroids(52).
Spheroids naturally mimic various aspects of solid tissues and are equipped with inherent gradients for efficient diffusion of oxygen and nutrients as well as the removal of metabolic wastes. These cellular aggregates can emulate avascular, solid tumor behavior more effectively than standard 2D environments because spheroids, much like tumors, usually contain a heterogeneous population of surface-exposed and deeply buried cells, proliferating and non-proliferating cells, and well-oxygenated and hypoxic cells(53). Additionally, differentiation of pluripotent stem cells (PSCs) typically involves formation of spherical structures called embryoid bodies, an important step for subsequent cell differentiation studies in vitro. Spheroids thus represent an especially good physiological 3D model for studying solid tumorigenesis and stem cell differentiation. In addition, spheroids can be readily analyzed by imaging using light, fluorescence, and confocal microscopy, which is an advantage over more complex 3D cell culture models. Furthermore, it is relatively simple to mass-produce uniformly sized 3D spheroids making them highly amenable for many in vitro high throughput and toxicity screening applications.
3D Cultures using Hydrogels and Extracellular Matrices
Hydrogels are comprised of networks of cross-linked polymer chains or complex protein molecules of natural or synthetic origin. Due to their significant water content, hydrogels possess biophysical characteristics very similar to natural tissue, and serve as highly effective matrices for 3D cell culture. Hydrogels can be used as stand-alone 3D matrices or combined with other technologies, such as solid scaffolds, permeable supports, cellular microarrays, and microfluidics devices.
Hydrogels can be employed in 3D culture systems in a variety of ways. They can be used as a coating reagent for various cell culture surfaces including solid scaffolds. Alternatively, cells can be encapsulated in or sandwiched between these matrices. The morphology, growth and functionality of cells within the hydrogel matrices depend on the presentation of biophysical and biochemical cues, as well as physical properties such as permeability and matrix stiffness.
Natural Hydrogels and Extracellular Matrices (ECMs)
Naturally derived hydrogels for cell culture are typically formed of proteins and ECM components such as collagen, laminin, fibrin, hyaluronic acid, chitosan, or Corning Matrigel® matrix. Derived from natural sources, these gels are inherently biocompatible and bioactive(54). They also promote many cellular functions due to the presence of various endogenous factors, which can be advantageous for supporting viability, proliferation, function, and development of many cell types(55). These materials are widely used for studying tissue and disease models (Table 1) and for a variety of 3D co-culture models (Table 2).
| Application | Cell Type | 3D Model | Culture Substrate and Matrix | Reference |
|---|---|---|---|---|
| Stem Cell Differentiation | Human PSCs | Differentiation to ureteric-bud committed renal progenitor cells | Corning® Matrigel® matrix | 94 |
| Human PSCs | Differentiation to cardiac microtissues | Aligned collagen I | 124 | |
| Human PSCs | Differentiation to Endoderm | Collagen I | 125 | |
| Human ESCs | Differentiation to hepatocytes | Collagen I scaffold; 2D collagen I-coated dish | 126 | |
| Human ESCs | Differentiation to smooth muscle, neurons, and hepatocyte-like cells | Polymer scaffold with ECM coating | 127 | |
| Human ESCs | Differentiation to neural precursors | Hyaluronic acid matrix | 128 | |
| Mouse ESCs | Differentiation to renal tubular cells | Corning Matrigel matrix; Ultra-Low Attachment plates | 93 | |
| Mouse ESCs | Differentiation to inner ear sensory epithelia | Corning Matrigel matrix | 92 | |
| Mouse ESCs | Differentiation to neurons | Collagen I scaffold | 129 | |
| Mouse ESCs | Differentiation to neurons and astrocytes | Fibrin scaffold | 130 | |
| Mouse ESCs | Differentiation to chondrocytes | PEG scaffold | 131 | |
| Human NPCs | Differentiation to neuronal cells | Corning PuraMatrix™ (RADA-16) scaffold | 132 | |
| Human NPCs | Differentiation to neuronal cells | Corning PuraMatrix ± functionalized matrix (laminin I) | 133 | |
| Mouse primary neuronal cells | Differentiation to osteogenic lineage cells in 2D and 3D systems | PLLA and polystyrene polymer scaffolds coated with Collagen I | 134 | |
| Human MSCs | Differentiation to osteocytes | Corning PuraMatrix peptide hydrogel | 135 | |
| Canine AdMSCs | Differentiation osteogenic lineage cells in 2D, 3D, and in vivo (implantation in dog) systems | Corning Matrigel matrix | 136 | |
| Human dental pulp stem cells | Differentiation to osteocytes | Corning Matrigel matrix; Collagen sponge | 137 | |
| Organoid Generation | Human PSCs | Intestinal organoids | Corning Matrigel matrix | 95, 96 |
| Mouse ESCs | Retinal optic-cup structure | Corning Matrigel matrix | 97 | |
| Mouse embryonic pancreatic progenitors | Pancreatic organoid | Corning Matrigel matrix | 98 | |
| Mouse epithelia | Mammary organoid | Corning Matrigel matrix | 138 | |
| Mouse primary colon tumor cells | Primary mouse colon tumor organoids | Corning Matrigel matrix | 139 | |
| Mouse ESCs | Functional thyroid follicular cells | Corning Matrigel matrix | 140 | |
| Human ESCs | Cerebral organoid model of human brain development and microcephaly | Corning Matrigel matrix | 141 | |
| Application | Cell Type | 3D Model | Culture Substrate and Matrix | Reference |
|---|---|---|---|---|
| Cancer Co-Culture Models | Mouse mammary tumor cells + endothelial cells | Co-culture of tumor spheroid to study impact of endothelial cells on tumor growth, vascularization and metastasis in vitro and in vivo | Hanging drop; implantation in nude mice | 111 |
| Primary breast epithelial cells + endothelial cells | Co-culture model to study the role of endothelial cells in growth of normal and cancerous breast epithelial cells in 3D cultures | Transwell® filters (0.4 µm); Corning® Matrigel® matrix | 112 | |
| Mesenchymal stem cells + ovarian cancer cells (OCC) | Co-culture model to study the influence of MSCs on OCC migration and invasion in an amniochorionic membrane model | Transwell® filters; Ultra-Low Attachment plates; Corning Matrigel matrix; Amniotic membrane scaffold | 114 | |
| Mouse breast cancer cell line + stromal fibroblasts | Co-culture model to study the effects of stromal cells on breast cancer progression | Corning Matrigel matrix | 109 | |
| Keratinocytes + melanocytes + dermal stem cells + melanoma cells | A 3D skin reconstruct model to study melanoma progression in human skin | Bovine Collagen I | 150 | |
| Human melanoma cells + keratinocytes | Organotypic skin melanoma spheroid model for in vitro drug testing | Collagen I | 151 | |
| Multi-Cellular Co-Culture Models | Human primary brain endothelial cells + human primary pericytes + human primary astrocytes | Blood-brain barrier model | Transwell inserts (0.4 µm) | 146 |
| Hepatocytes + mouse fibroblasts or bovine endothelial cells | Micropatterned co-culture of hepatocyte spheroids | Micropatterned PEG hydrogel; gelatin | 113 | |
| Human bronchial epithelial cells + endothelial cells | A model to recapitulate lung morphogenesis | Transwell inserts (0.4 µm); Corning Matrigel matrix | 42 | |
| Primary rat hepatocytes + rat liver endothelial cells | A physiologically relevant co-culture model of hepatic sinusoids | Collagen I | 147 | |
| Alveolar cell line + macrophage-like cells + mast cells + endothelial cells | Tetraculture model mimicking alveolar barrier to study toxic effects of particles on the lungs; cells cultured at air-liquid interface (ALI) | Transwell inserts (0.4 µm) | 39 | |
| Mesenchymal stem cells + alveolar epithelial cells | Novel collagen-drop cell migration assay for wound repair model | Transwell insert (3 µm); Collagen I | 148 | |
| Human retinal progenitor cells + endothelial cells | Hypoxia induced retinal neovascularization with and without endothelial cells | Transwell inserts (0.4 µm); Corning Matrigel matrix | 41 | |
| Human mammary epithelial cells + human fibroblasts + adipocytes | A physiologically relevant tri-culture system to model human breast tissue | Corning Matrigel matrix + Collagen I; porous silk protein scaffold | 149 | |
| Human bronchial epithelial cells | Differentiation of cells to glandular acini on ALI | Transwell Collagen IV-coated; Corning Matrigel matrix | 152 | |
| Polarized airway epithelial cells | Infection of polarized epithelial cells from normal and cystic fibrosis patients. Cells were grown at ALI. | Transwell inserts (0.4 µm) | 40 | |
The ECM provides a complex, nanoscale architecture of structural proteins such as collagen, laminin, and fibronectin to create the mechanical properties inherent in the cellular microenvironment(90,91). Cells sense these mechanics through their cell surface integrins, and bind to specific adhesion motifs present on the ECM proteins. The ECM is vital for sequestering soluble biomolecules and growth factors, and releasing these signaling molecules with spatial-temporal control to guide processes such as cell migration, matrix degradation and deposition(90,91). ECM remodeling is imperative for achieving tissue homeostasis and is particularly pronounced during development and diseases. Thus, to truly mimic the ECM in vivo, it is necessary to develop 3D culture models that exhibit the mechanical and chemical properties of the ECM, not only at the initial stage of cell seeding, but rather, in a dynamic and tunable manner as the cells grow and develop.
Corning® Matrigel® matrix is an ECM-based natural hydrogel that has been used extensively for 3D cell cultures in vitro and in vivo. This reconstituted basement membrane is extracted from Engelbreth-Holm-Swarm (EHS) mouse tumors and contains all of the common ECM molecules present within basement membrane (i.e., laminin, collagen IV, heparin sulfate proteoglycan, and nidogen/entactin)(56). The ECM components of Matrigel activate cancer cell signaling pathways that regulate angiogenesis(57,58), tumor cell motility(59), and sensitivity to anti-tumor drugs(60). Since Matrigel mimics the in vivo basement membrane, it is often used for studying cancer cells that resemble cells that reside in epithelial tissues(61). Dr. Mina Bissell’s pioneering research on mammary epithelial cells and breast cancer has utilized Matrigel matrix, or an equivalent, which demonstrated the enabling power of 3D cell culture for creating in vivo-like model systems(62) and the importance of integrin signaling in cancer(63,64).
Endothelial cells line the closed internal body cavities (e.g., blood vessels, bile ducts) and play an integral role during angiogenesis. The primary stages of angiogenesis include endothelial cell migration and invasion through the vascular basement membrane, and the subsequent process of tubulogenesis (tube formation). To study endothelial cell tubulogenesis, many laboratories utilize 3D cell culture environments comprised of Corning Matrigel Matrix to promote endothelial cell differentiation and the formation of capillary-like structures in vitro (Figure 2).

Collagen Type I is another commonly used natural hydrogel that is used for 3D cell culture. Collagen I can be isolated from various biological sources including bovine skin, rat tail tendon, and human placenta. This ECM molecule can also be electrospun into membranes(65,66), and can support 3D cell growth and differentiation. Additionally, Collagen I interacts with integrin receptors to modulate gene expression(67). The target genes modulate the production of matrix metalloproteinases (MMPs), the enzymes that degrade ECM components during tumor metastasis(68), as well as genes that affect cell sensitivity to drugs(69) and cell migratory function(70,71).
Synthetic Hydrogels
Synthetic hydrogels are a good choice for 3D cell culture applications when naturally derived biological matrices are unsuitable. Synthetic hydrogels are comprised of purely non-natural molecules (reviewed in 75) such as poly(ethylene glycol) (PEG)(76), poly(vinyl alcohol)(77), and poly(2-hydroxy ethyl methacrylate)(78). They are biologically inert, but provide structural support for various cell types. PEG hydrogels have been shown to maintain the viability of encapsulated cells while allowing for ECM deposition as the hydrogel degrades(79), thereby demonstrating that synthetic gels can function as 3D cell culture platforms in the absence of integrin-binding ligands.
It is also possible to modify inert synthetic hydrogels with appropriate biological components. An example of a synthetic hydrogel that is tunable is Corning PuraMatrix™ peptide hydrogel, a self- assembling, synthetic oligopeptide that exhibits nanometer scale fibers. Nanometer-sized fibers and pores are essential to ensure a true 3D environment for the cell(80,81). An additional advantage of PuraMatrix peptide hydrogel is the ability to customize the material with specific peptide sequences to improve cell attachment, cell homing, and other behaviors(82). To achieve optimal cell growth and differentiation using PuraMatrix peptide hydrogel, it is necessary to supplement the hydrogel with appropriate bioactive molecules (e.g., growth factors, ECM proteins, and/or other molecules). Functionalization with appropriate biological factors can convert inert synthetic matrices such as Corning® PuraMatrix™ peptide hydrogel into useful in vitro models for supporting differentiation of stem and progenitor cells. As this field advances, there will be a need for matrices with combined properties of natural and synthetic hydrogels.
Future Directions
In vitro 3D cell culture models have become increasingly sophisticated and widely used to study cell growth, tissue morphogenesis, stem cell differentiation, disease modeling, and drug discovery. It is also evident that different 3D models with varying characteristics are required to meet the needs of specific cell types or applications. To date, many complex 3D models of tumorigenesis, stem cell differentiation, and organoid formation make use of natural ECM-based hydrogels, such as Corning® Matrigel® matrix or collagen. Combining newer technologies such as microfluidics or bio-printing with biologically relevant materials, such as Matrigel matrix, may help to provide novel systems and models for cell biology research.
About the Authors
Suparna Sanyal, Ph.D. is a freelance science writer and cell biologist with extensive knowledge of angiogenesis and stem cell biology. Marshall Kosovsky, Ph.D. is the Global Technical Support Manager at Corning Life Sciences (kosovskym@corning.com).
Links to more information about 3D cell culture:
Click here for a link to the full review article on 3D Cell Culture
Click on the following for review articles focusing on Corning materials and systems for optimizing the culture of stem cells:
References
Note: Some references have been omitted from this blog post for brevity. To view all the references, click on the “Full Review Article,” link.
1. Lodish, H. et al. Molecular Cell Biology. New York: W.H. Freeman and Company, (2002).
2. Kleinman, H. K. et al. Curr. Opin. Biotech. 14:526-532 (2003).
3. Bissell, M. J. et al. Differentiation 70:537-546 (2002).
8. Pampaloni, F. et al. Nat. Rev. Mol. Cell Biol. 8:839-845 (2007).
9. Lee, J. et al. Tissue Eng. 14(1):61-86 (2008).
10. Haycock, J. W. Methods Mol. Biol. 2011; 695:1-15 (2011).
11. 3D Cell Culture Trends 2010 Report, published by HTStec Limited, Cambridge, UK, February 2010.
13. Baker, B.M. and Chen, C.S. J. Cell Sci. 125(13):3015-3024 (2012).
14. Kola, I. Clin. Pharmacol. Ther. 83(2):227-230 (2008).
15. Singh, S. S. Curr, Drug Metab. 7(2):165-182 (2006).
18. Khetan, S. and Burdick, J. A. Biomaterials 31:8228-8234 (2010).
19. Singhvi, R. et al. Science 264:696-698 (1994).
20. Chen, C. S. et al. Science 276:1425-1428 (1997).
21. Thomas, C. H. et al. Proc. Natl. Acad. Sci. USA 99:1972-1977 (2002).
22. McBeath, R. et al. Dev. Cell 6:483-495 (2004).
23. von der Mark, K. et al. Nature 267:531-532 (1977).
24. Petersen, O. W. et al. Proc. Natl. Acad. Sci. USA 89:9064-9068 (1992).
25. Benya, P. D. and Shaffer, J. D. Cell 30:215-224 (1982).
26. Emerman, J. T. and Pitelka, D. R. In Vitro 13:316-328 (1977).
27. Lee, E. Y. et al. J. Cell Biol. 98:146-155 (1984).
28. Brock, A. et al. Langmuir 19:1611-1617 (2003).
29. The Théry, M. et al. Proc. Natl. Acad. Sci. USA 103:19771-19776 (2006).
30. The Théry, M. et al. Nature 447:493-496 (2007).
31. Mahmud, G. et al. Nat. Phys. 5:606-612 (2009).
32. Kilian, K. A. et al. Proc. Natl. Acad. Sci. USA 107:4872-4877 (2010).
33. Debnath, J. and Brugge, J. S. Nat. Rev. Cancer 5:675-688 (2005).
34. Nelson, C. M. and Bissell, M. J. Annu. Rev. Cell Dev. Biol. 22:287-309 (2006).
35. Yamada, K. M. and Cukierman, E. Cell 130:601-610 (2007).
36. Cukierman, E. et al. Science 294:1708-1712 (2001).
37. Grinnell, F. Trends Cell Biol. 13:264-269 (2003).
38. Walpita, D. and Hay, E. Nat. Rev. Mol. Cell Biol. 3:137-141 (2002).
39. Klein, S. G. et al. Particle and Fibre Tox. 10:31-47 (2013).
40. Mitchell, G. et al. Infection and Immunity 79(9):3542-3551 (2011).
41. Kumar, R. et al. Vascular Cell, 3:27-41 (2011).
42. Franzdóttir, S. R. et al. Respir. Res. 11:162-171 (2010).
43. Birgersdotter, A. et al. Semin. Cancer Biol. 15:405-412 (2005).
44. Li, C. et al. Cancer Res. 66:1990-1999 (2006).
45. Ghosh, S. et al. J. Cell Physiol. 204:522-531 (2005).
46. Delcommenne, M. and Streuli, C. H. J. Biol. Chem. 270:26794-26801 (1995).
47. Fuchs, E. et al. Cell 116:769 (2004).
48. Gómez-Lechón, M. J. et al. J. Cell Physiol. 177:553-562 (1998).
49. Berthiaume, F. et al. FASEB J. 10:1471-1484 (1996).
50. Semino, C. E. et al. Differentiation 71:262-270 (2003).
51. Powers, M. J. et al. Tissue Eng. 8:499-513 (2002).
52. Mueller-Klieser, M. et al. Br. J. Cancer 53:345-353 (1986).
53. Frieboes, H.B. et al. Cancer Res. 66:1597-1604 (2006).
54. Dawson E. et al. Adv. Drug. Deliv. Rev. 60(2):215-228 (2008).
55. Tibbitt, M. W. and Anseth, K. S. Biotechnol. Bioeng. 103(4):655 (2009).
56. Kleinman, H. K. and Martin G. R. Semin Cancer Biol. 15:378-386 (2005).
57. Languino, L. R. et al. J Cell Biol. 109:2455-2462 (1989).
58. Zhou, Z. et al. Cancer Res. 64:4699-4702 (2004).
59. Carpenter, P. M. et al. Mol. Cancer Res. 7:462-475 (2009).
60. Miyamoto, H. et al. Pancreas 28:38-44 (2004).
61. Gurski, L. A. et al. Oncology Issues 25:20-25 (2010).
62. Barcellos-Hoff, M. H. Development 105:223-235 (1989).
63. Weaver, V.M. J. Cell Biol. 137:231-245 (1997).
64. Bissell, M. J. et al. Curr. Opin. Cell Biol. 15:753-762 (2003).
65. Sisson, K. et al. Biomacromolecules 10(7):1675-1680 (2009).
66. Hartman, O. et al. Biomacromolecules 10(8):2019-2032 (2009).
67. Kiefer, J. A. and Farach-Carson, M. C. Matrix Biol. 20:429-437 (2001).
68. Ellerbroek, S. M. et al. J. Biol. Chem. 276:24833-24842 (2001).
69. Kim, Y. J. et al. Int. J. Biol. Macromol. 45:65-71 (2009).
70. Menke, A. et al. Cancer Res. 61:3508-3517 (2001).
71. Hall, C. L. et al. Neoplasia. (New York, NY) 10:797-803 (2008).
75. DeVolder, R. and Kong, H. J. Wiley Interdiscip. Rev. Syst. Biol. Med. 4(4):351-365 (2012).
76. Sawhney, A. S. et al. Macromolecules 26(4):581-587 (1993).
77. Martens, P. and Anseth K. S. Polymer 41(21): 7715-7722 (2000).
78. Chirila, T. V. et al. Biomaterials 14(1):26-38 (1993).
79. Bryant, S. J. and Anseth, K. S. J. Biomed. Mater. Res. 59(1):63-72 (2002).
80. Gelain, F. et al. PLoS ONE 1: e119 (2006).
81. Horii, A. et al. PLoS ONE 2(2): e190 (2007).
82. Zhang, S. Nature Biotech. 22:151-152 (2004).
92. Koehler, K.R. et al. Nature 500:217-221 (2013).
93. Morizane, R. et al. PLoS One 8(6):e64843 (2013).
94. Xia, Y. et al. Nat. Cell Biol. 15(12):1507-1515 (2013).
95. Spence, J. R. et al. Nature 470:105-109 (2011).
96. McCracken, K. W. et al. Nat. Protoc. 6(12):1920-1928 (2011).
97. Eiraku, M. et al. Nature 472:51-56 (2011).
98. Greggio, C. et al. Development 140:4452-4462 (2013).
99. Gray, R. S. et al. Curr. Opin. Cell Biol. 22(5): 640-650 (2010).
109. Li, L. and Lu, Y. J. Cancer 2:458-466 (2011).
111. Upreti, M. et al. Transl. Oncol. 4(6):365-376 (2011).
112. Ingthorsson, S. et al. BMC Res. Notes 3:184 (2010).
113. Otsuka, H. et al. Sci. Technol. Adv. Mater. 14:065003 (2013).
114. Touboul, C. et al. J. Transl. Med. 11:28 (2013).
124. Thavandiran, N. et al. Proc. Natl. Acad. Sci. USA 110(49):E4698-4707 (2013).
125. Turovets, N. et al. Cell Transplantation 21:217-234 (2012).
126. Baharvand, H. et al. Int. J. Dev. Biol. 50(7):645-652 (2006).
127. Carlson, A. L. et al. FASEB J. 26:3240-3251 (2012).
128. Brannvall, K. et al. J. Neurosci. Res. 85:2138-2146 (2007).
129. Li, X. et al. Prog. Polymer Sci. 37(8):1105-1129 (2012).
130. Willerth, S. M. et al. Biomaterials 27:5990-6003 (2006).
131. Hwang, N. S. et al. Stem Cells 24:284-291 (2006).
132. Liedmann, A. et al. Biores. Open Access. 1(1):16-24 (2012).
133. Ortinau, S. et al. Biomed. Eng. Online 9:70-87 (2010).
134. Lai, Y. et al. PLoS One 7(9):e45074 (2012).
135. Chen, J. et al. Tissue Eng. Part A. 19(5-6):716-728 (2013).
136. Kang, B.-J. et al. J. Vet. Med. Sci. 74(7):827-836 (2012).
137. Riccio, M. et al. Eur. J. Histochem. 54(4):e46 (2010).
138. Ewald, A. J. et al. Dev. Cell. 14(4):570-581 (2008).
139. Xue, X. and Shah, Y. M. J. Vis. Exp. 75:e50210 (2013).
140. Antonica, F., et al. Nature 49:66-71 (2012).
141. Lancaster, M. A. et al. Nature 501:373-379 (2013).
146. Urich, E. et al. Sci. Rep. 3:1500 (2013).
147. Kim, Y. and Rajagopalan, P. PLoS One 5(11):e15456 (2010).
148. Akram, K. M. et al. Resp. Res. 14:9 (2013).
149. Wang, X. et al. Biomaterials 31(14):3920-3929 (2010).
150. Li, L. et al. J. Vis. Exp. 54:e2937 (2011).
151. Vörsmann, H. et al. Cell Death Dis. 4(7): e719 (2013).
152. Wu, X. et al. Am. J. Respir. Cell Mol. Biol. 44:914-921 (2011).