We are seeing very poor DNA yield. Do you have a good protocol?
This question is part of the following Ask The Expert session:
Plasmid DNA Production for Cell and Gene Therapy
Answered by:
Company: MilliporeSigma
Job Title: Associate Director, EMEA Segment Development for Cell and Gene Therapy
Answer
In order to have a good plasmid DNA (pDNA) yield, the best protocol is to have a robust overall process where each step is well optimized. The pDNA purification is challenging due to its quite large size, high negative charge, viscosity and the fact that the contaminants have similar properties to the pDNA (open circular pDNA, genomic DNA, high molecular weight RNA). In addition, large plasmids can be sensitive to shear stress, further complicating the purification. In order to purify the supercoiled pDNA (desired form for therapeutics/transfection) with high yield, a well optimized downstream process is needed. Our company has solutions and capabilities for each step of the downstream process to assure an optimum pDNA yield and purity (summarized in figure 1). Considerations and watch outs for each downstream unit operations are discussed below.
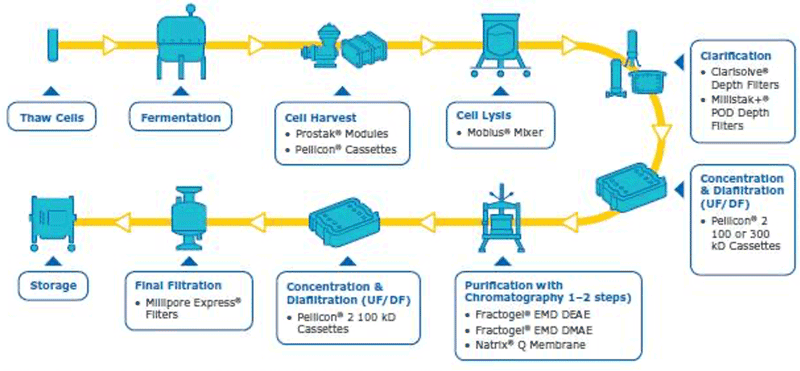
Cell harvest
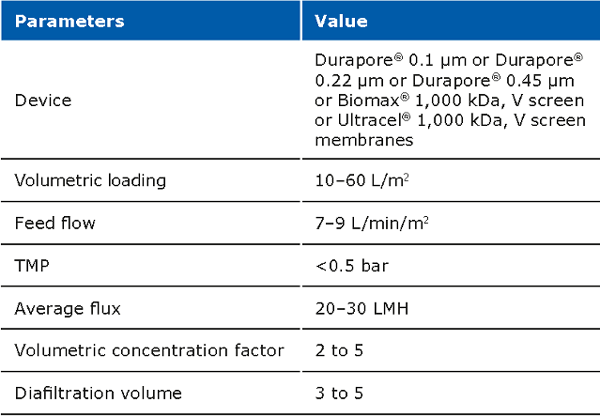
The cell harvest step uses centrifugation or microfiltration TFF. Centrifugation is often more cost effective when batch volumes (<10 L) or larger batch volumes (>1,000 L) need to be processed. When using centrifugation, special attention is needed for high shear generated in large scale centrifugation. Microfiltration can be performed with open-channel, flat-sheet TFF devices such as ProstakTM cassettes with Durapore® 0.1 or 0.2 μm microfiltration membranes or Pellicon® cassettes with Durapore® V screen or Biomax® 1,000 kD V screen ultrafiltration (UF) membranes. The open feed channels create a gentle flow path for cell retention, cause low shear, and can be used to process viscous and/or high solid feeds. When using membrane cut-offs such as these, it is important to utilize a two-pump (permeate-controlled) TFF system. The TFF harvest step typically involves a 2-5X volumetric concentration followed by a 3–5 volume diafiltration for washing out spent media components and extracellular impurities prior to further downstream purification. TFF harvest is typically operated at low transmembrane pressure (TMP; 3–5 psi) and ΔP (<7 psi) with a control on the permeate flux. Hollow fiber modules are also applicable, but may be accompanied with scaling issues due to the non-linear scalability.
Typical operating parameters for micro filtration TFF:
Cell Lysis
The methods used for cell lysis can be divided into two main categories – chemical (alkali, detergents, enzymes, osmotic shock) and physio mechanical (heat, shear, agitation, ultra-sonification, and freeze-thawing) lysis. Alkaline lysis (NaOH at pH ~12) accompanied by detergents such as sodium dodecyl sulfate (SDS) and Triton® X-100 is the most common approach. It is important to optimize the lysis incubation time as it directly impacts the quality and quantity of plasmid DNA. Longer incubation time could lead to irreversible denaturation of plasmid DNA and shear degradation of genomic DNA. It is critical to have efficient but not too aggressive mixing employed on the alkaline lysis step to ensure there are no pH extremes causing irreversible denaturing of the plasmids or degrading it due to excessive shear. Mobius® single-use mixers can be very effective for batch lysis.
During the alkaline lysis method, cells are treated at specific, narrow range of pH (typically around pH 12) at which the genomic DNA will be irreversibly denatured, while the pDNA double chain remains intact (pH range of 12.0 to 12.5). The optimum pH value varies depending on the type of plasmid and host strain. A deviation of more than 0.1 pH unit from the optimum value may affect the yield and it is therefore critical to maintain a tight control of the pH range during alkaline lysis. At a pH >12.5, pDNA becomes irreversibly denatured and if the pH is too low, genomic DNA won’t be completely denatured and could complicate further downstream purification process. The incubation time for a standard alkaline lysis is fairly short and the step is usually completed typically within 5 minutes.
After the alkaline lysis, the pH is neutralized. This is typically carried out by addition of a high-salt buffer, such as sodium or potassium acetate at concentration of 0.7M – 3.0M and pH ~5–7.5, with/without 1.0–1.5% CaCl2 (promotes RNA precipitation) in the presence of a detergent (0.2 – 1% SDS). Polyethylene glycol (PEG) and polyethylenimine can also be added to the neutralization buffer to promote the precipitation of genomic DNA. Homogenous mixing during neutralization and precipitation is critical to maintain pDNA quality.
Clarification
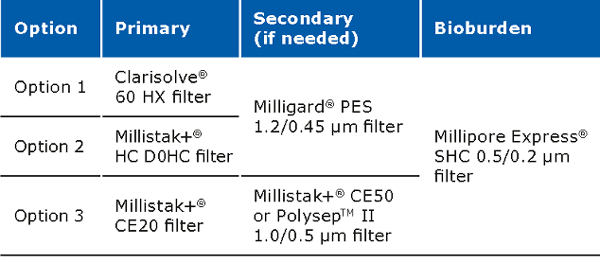
Clarification unit operations for pDNA processes should enable removal of solid content from the feed stream. Clarification can be performed via centrifugation and/or normal flow filtration. Traditionally, centrifugation is used for the clarification. When using centrifugation, one should pay attention to the shear stress, which may damage the structure of the supercoiled plasmid, resulting in lower yields. Centrifugation may also require a secondary clarification step. Clarification using normal flow filtration is the modern approach. When using this method, feed streams can be used which are either untreated, pretreated or preclarified. Pretreatment has a major impact on the clarification filter capacity and must be selected carefully along with a consideration of the scalability of the process. Pretreatment options include use of gravity settling and separation, Polygard® CR 1 µm/Polygard® CR 50 µm, bag filters stainless steel screen filters, paper filters, and centrifugation. The capacity of the Polygard® CR filters is typically in the range of 0.55–8 L/inch.
The recommended filter set ups for the clarification step are:
The expected capacity of the filters is strongly dependent on whether a pretreatment is performed:
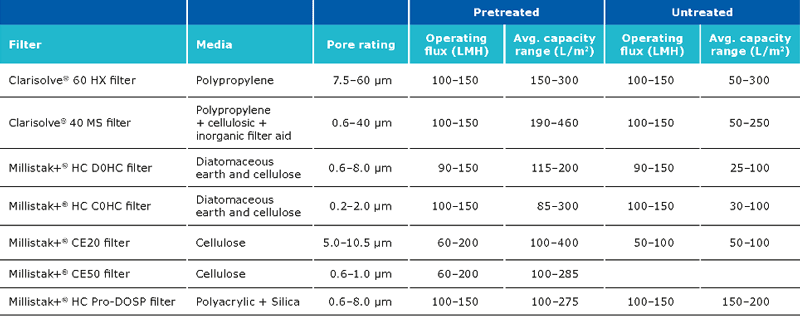
The typical recovery when using Clarisolve® and Millistak+® filters is >90%. The recovery can be increased using a chase wit salt containing buffer. The capacity of the Millipore Express® SHC sterilizing grade filter post clarification is generally between 400-650 L/m2.
Tangential flow filtration (TFF)
After clarification, there can be opted for a TFF step. This will allow to concentrate and perform diafiltration to exchange the medium into a buffer suitable for the downstream chromatography step. By performing a concentration prior to chromatography, one can reduce the chromatography loading time. In addition, RNA, small sized genomic DNA, and small proteins can be removed during TFF, which also helps prevent fouling of chromatography resins.
When performing TFF, some key watch outs are:
- Shear sensitivity of plasmids, careful adjustment of process parameters is needed to prevent damage to the plasmid.
- Membrane fouling, which results into yield loss.
- High salt buffers promote plasmid compaction, which may allow passage through small pores, resulting in yield loss.
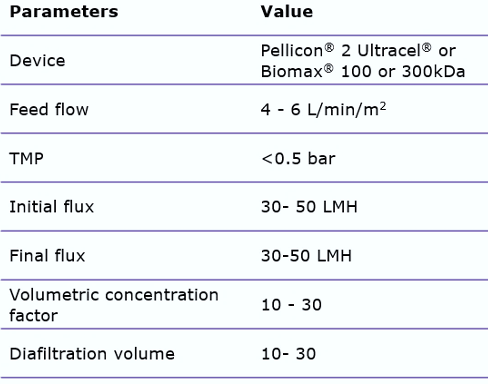
These issues can be avoided by carefully screening recommended open channel membranes, such as the Pellicon® 2 Ultracel® or Biomax® 100 or 300 kDa C-screen. By using open channels, shear stress can be reduced. In addition, minimizing the process time by appropriate membrane sizing is important. When using these wider membrane cut-offs, it is important to control the permeate flux, by utilizing a two-pump (permeate-controlled) TFF system. This will help prevent membrane fouling.
Typical TFF process parameters are:
Chromatography purification
For chromatography purification, typically anion exchange chromatography (AEC) and hydrophobic interaction chromatography (HIC) are used. Both techniques have been implemented for capture or intermediate purification/polishing and are often combined.
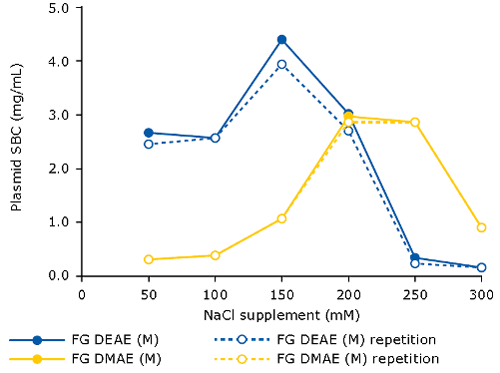
When using AEX as capture step for pDNA, it is beneficial to spike the clarified lysate with NaCl (120-250 mM). This will eliminate the interference of RNA, therefore increasing the binding capacity for pDNA. The optimal salt concentration for supplementation needs to be pre-determined, for example via a batch assay in microtiter plate format, measuring plasmid binding capacity at increasing sodium chloride concentrations. One can do this for different types of resins/membrane adsorbers. The following graph demonstrates this principle for the Fractogel® EMD DEAE (M) and Fractogel® EMD DMAE (M) resins:
Recommended resins for plasmid purification and their respective performance:
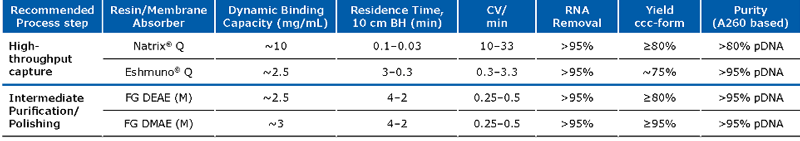
Due to the large size of the plasmid, typically low binding capacities are observed on commercially available resins. This is due to the fact that most AEX media were originally designed for protein purification, as plasmid molecules are much bigger than proteins, they cannot enter the small pores, causing low binding capacity and slow mass transfer. Because the Fractogel® and Eshmuno® resins contain tentacle technology, typically higher binding capacities compared to other available resins can be achieved. The Natrix® Q is based on membrane technology and contains large convective pores. The latter promotes mass transfer and improves binding capacities (5-10 mg/mL). Due to the membrane technology, very short residence times are needed (<0.2 min). Because of these advantages, together with its single use format, it has been widely adopted.
All four resins mentioned in the above table can be used for the capture of pDNA. However, only the Fractogel® EMD DEAE (M) and Fractogel® EMD DMAE (M) resins are well suited for intermediate purification or polishing of plasmid DNA due to their moderate binding capacity and flow as well as good resolution due to a medium bead size (d50: 48–60 μm), clearing residual impurities like RNA and endotoxin efficiently.
Examples of HIC resins are Capto™ PlasmidSelect and Toyopearl® Butyl. The HIC step can either be located before or after the AEX step. When the HIC step is the first chromatography step, the eluate can directly be processed onto Fractogel® resins, as pDNA binding to these AEX resins tolerate the presence of elevated concentration of ammonium sulfate (pDNA binding capacity is not negatively affected).
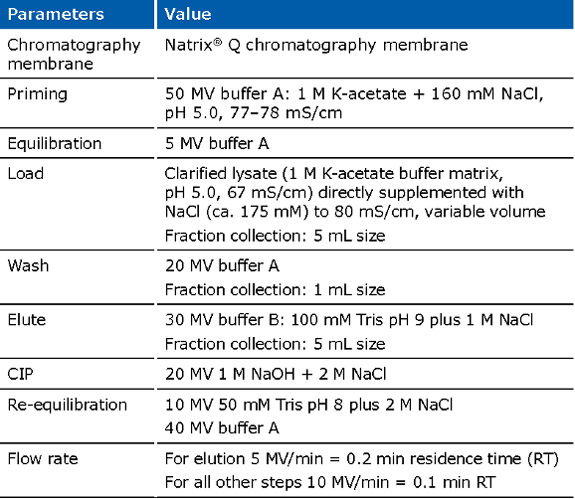
Example of an experimental run for the Natrix® Q chromatography membrane for capture of pDNA:
Final concentration and sterilizing grade filtration
After chromatography, the sample is concentrated and diafiltered in the buffer of choice. The same conditions apply as mentioned in the tangential flow filtration section.
The sterile filtration of pDNA can be challenging due to the large size and viscosity of the final product. In addition, large plasmids can be shear sensitive and may be damaged during the operation. Some key considerations are:
- Salt concentration: plasmid DNA tends to be more compact when increasing the salt concentration. The latter can lead to an improved yield and filter capacity.
- Membrane type: typically, a higher filter capacity and flux are achieved when using PES based membranes. PES based membranes can also be less damaging to larger plasmids (less shear). The Millipore Express® SHC is recommended for this operation.
- Plasmid purity: supercoiled plasmids tend to give better filtration performance compared to open-circular plasmids. Therefore, both yield and filter capacity tend to increase with purity.
- Defining the filtration endpoint: finding suggest that membrane fouling is correlated to yield loss. Optimizing the filtration endpoint can lead to an increase in yield.
- Feed flux or pressure do not show a significant impact on filtration capacity or yield. However, high driving forces should be avoided to lower potential shear stress.
Expected performance for the sterilizing grade filtration of purified pDNA:
