Using Electron Activated Dissociation for Comprehensive Characterization of O-linked Glycosylation
Glycosylation, a common post-translational modification (PTM), is crucial for antibody effector functions. Comprehensive characterization of N- and O-linked glycosylation in protein therapeutics is vital for ensuring drug safety and efficacy.
The industry standard approach for characterization of glycosylation via mass spec utilizes the MS/MS method Collision Induced Dissociation (CID). However, when CID is applied to glycopeptides, it can result in the loss of labile glycan moieties. This makes accurate determination of glycosylation sites challenging, especially for O-glycosylation without a consensus sequence. The alternative fragmentation method electron activated dissociation (EAD) extends beyond CID in glycopeptide analysis as it preserves glycan structures in the fragments, facilitating accurate localization. A single injection workflow combining both methods enables a more comprehensive characterization profile.
A recent technical note “Comprehensive characterization of O-linked glycosylation in etanercept by electron activated dissociation (EAD),” illustrates the importance and effectiveness of EAD to identify and locate O-linked glycosylation accurately. The work of this current tech note was done using Etanercept, a dimeric fusion protein consisting of two tumor necrosis factor receptor (TNFR)-Fc chains with 3 N-glycosylation and 13 O-glycosylation sites on each chain. EAD’s ability to identify glycosylation positions facilitated the distinction between positional isomers of O-glycopeptides. While this work was done on etanercept, the EAD-based workflow described in this technical note could be applied to elucidate complex glycosylation profiles in other protein therapeutics.
This workflow uses the ZenoTOF 7600 system and Biologics Explorer software from SCIEX.
Technical Note Highlights
EAD accurately identified localization of O-glycosylation in etanercept
EAD data enabled localization of O-glycopeptides containing up to 7 O-glycans. Labile glycans can be retained in the EAD fragments, allowing unambiguous determination of glycosylation sites and confident differentiation of positional isomers of O-glycopeptides (Figure 1).
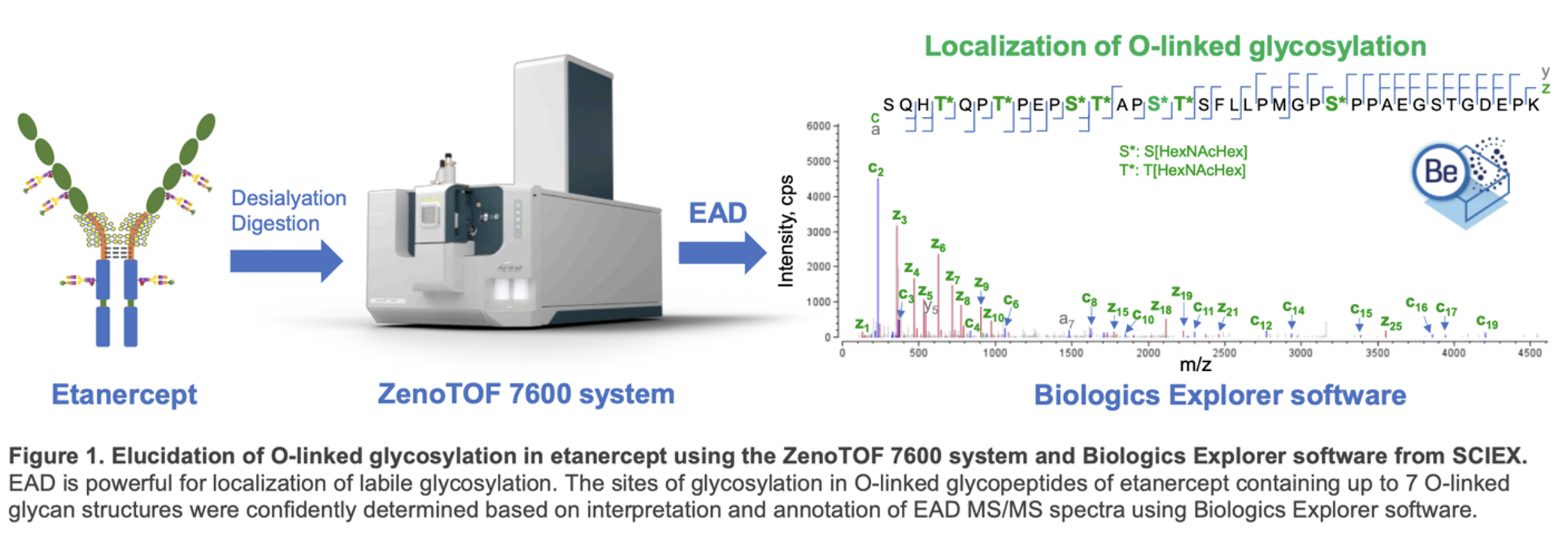
Glycosylation in etanercept consists of 3 N-glycosylation and 13 O-glycosylation sites on each TNFR-Fc chain. Most of the O-glycosylation sites are located in the hinge region. Due to loss of labile glycans, traditional collision-based MS/MS methods like CID struggle to provide precise site-specific glycosylation information.
EAD preserves glycans in fragments, enabling clear identification of this labile post-translational modification. This technical note utilized the EAD DDA or MRMHR method to analyze the intricate O-glycosylation pattern of desialylated etanercept.
The ZenoTOF 7600 system provides excellent sensitivity and high quality MS/MS
The Zeno trap of the ZenoTOF 7600 system provides a 5–10-fold increase in the detection of MS/MS fragments, leading to excellent MS/MS sensitivity and spectral quality.
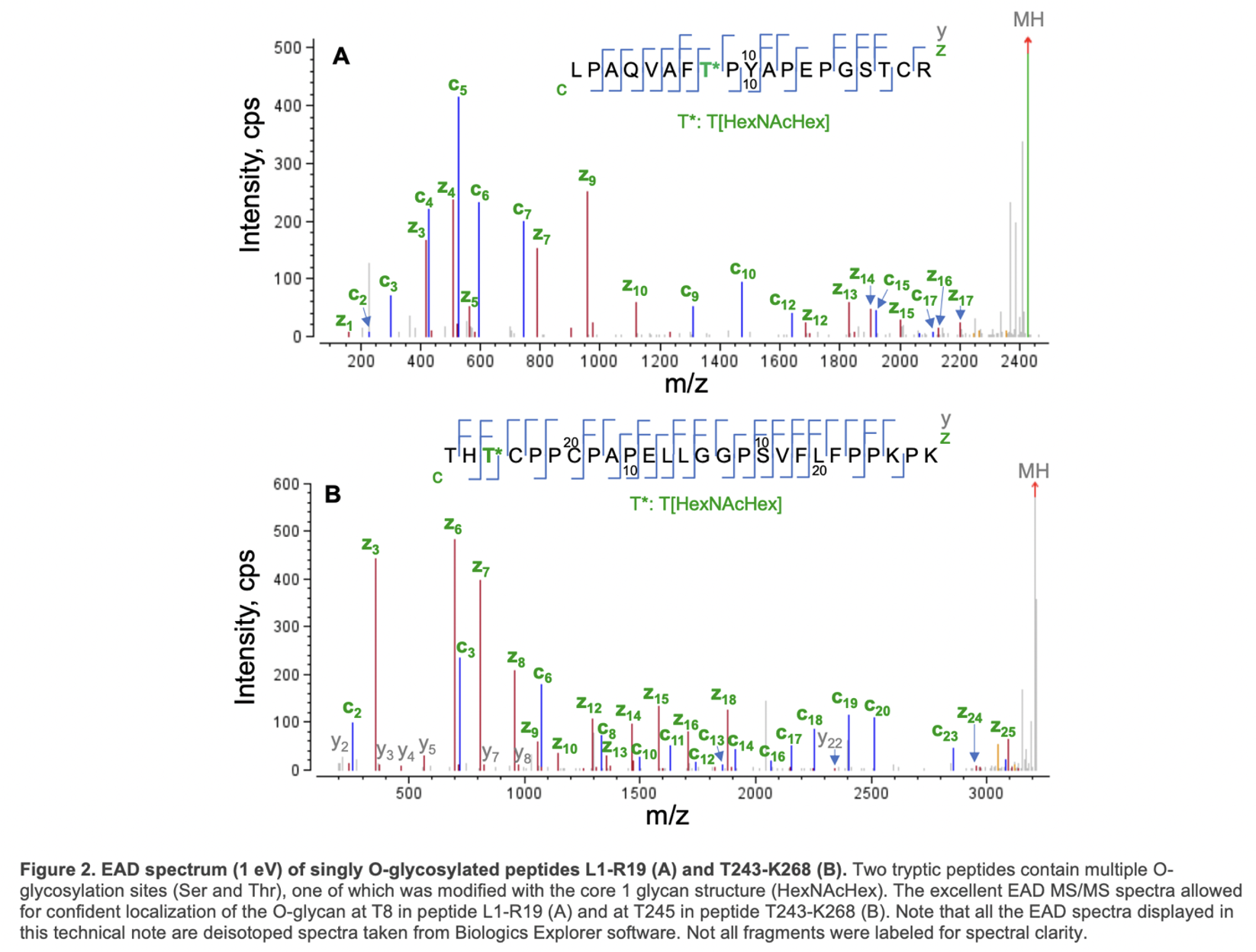
EAD generated an MS/MS spectra with a nearly complete series of c/y/z fragments, leading to confident identification of these two O-glycopeptides and unambiguous localization of the glycan. Specifically, the O-glycosylation site in peptide L1-R19 was determined to be T8 based on the m/z of c7/c9 and y11/z12 fragments, while T245 in peptide T243-268 was modified with HexNAcHex based on the detection of the non-glycosylated c2/y23 and glycan-containing c3/y24/z24 (Figure 2).
EAD provides confident identification of O-glycopeptides, accurate localization of glycosylation and isomer differentiation in one experiment
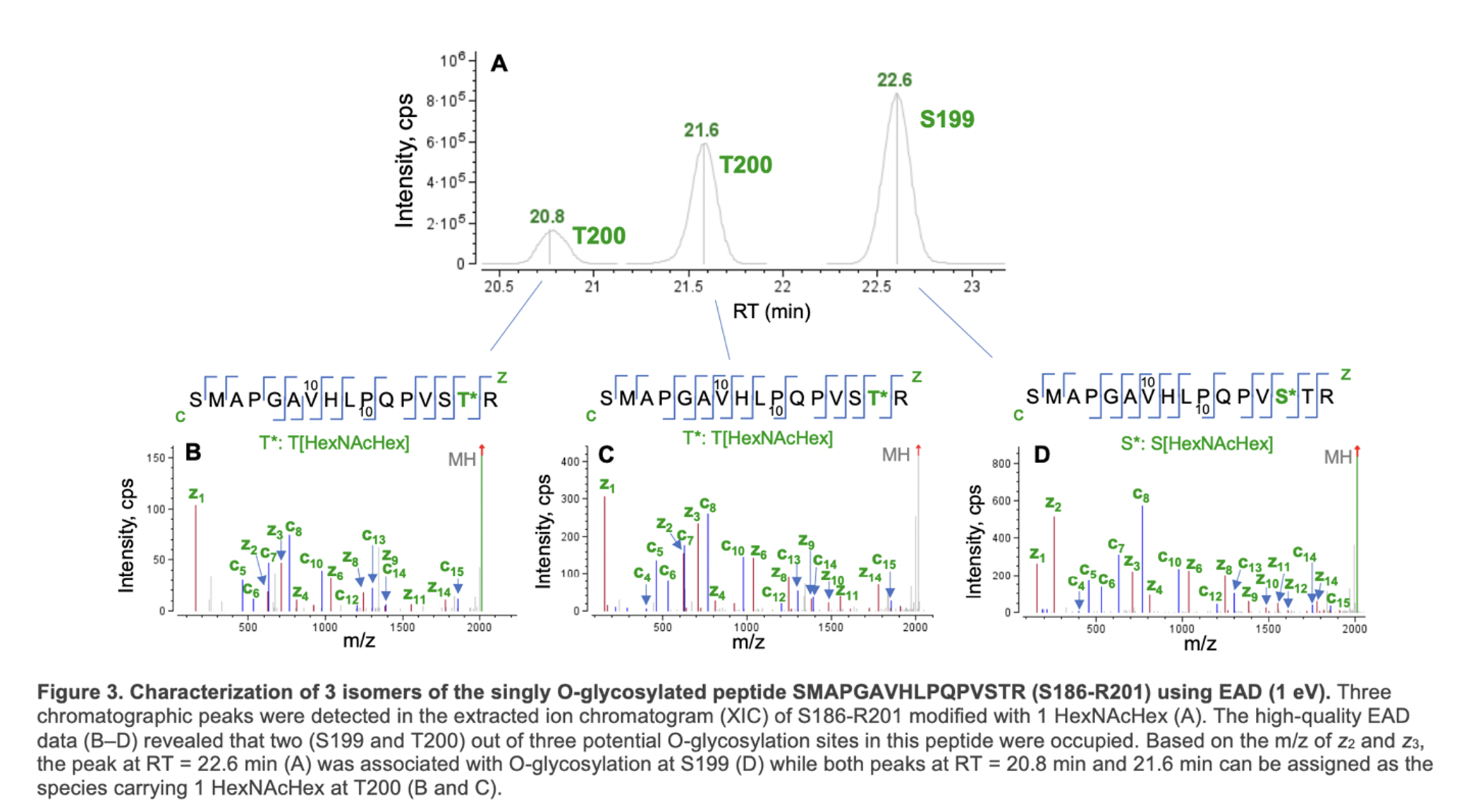
Figure 3 shows an example of EAD for the differentiation of positional isomers of a singly O-glycosylated peptide (S186- R201). Three isomeric species were observed in the XIC of O-glycopeptide S186-R201 (Figure 3A). The high-quality EAD data revealed that the most abundant isomer detected at RT = 22.6 min corresponded to O-glycosylation at S199 (Figure 3D) while the other two species (RT = 20.8 min and 21.6 min) were modified with 1 HexNAcHex at T200 (Figure 3B and 3C). The presence of two T200 isomers might be attributed to different linkages of the glycan. This result highlights the unique capabilities of EAD for confident identification of O-glycopeptides, accurate localization of glycosylation and isomer differentiation in one experiment.
EAD offers unparalleled power for unambiguous differentiation of positional isomers
The EAD MS/MS spectra illustrate the glycosylation patterns of O-glycopeptide S186-R201 of etanercept. For doubly glycosylated species, precise localization of 2 HexNAcHex at S199 and T200 was achieved by detecting c/z fragments with or without the glycan. Triple glycosylated species confirmed the presence of HexHAcHex at the N-terminus Ser residue, as expected. Despite the presence of 11 potential O-glycosylation sites in the S202-K238 peptide, EAD data revealed glycosylation at 7 of these sites. Multiple proline residues limited the generation of traditional c/z fragments, but additional a/y fragments allowed confident localization of 6 or 7 HexNAcHex. Two positional isomers were observed for glycopeptide S202-K238 with 6 HexNAcHex moieties, distinguished by signature EAD fragments such as doubly charged c11, c12, and c15. The m/z difference in c12 indicated glycosylation at T213 in Isomer 1 and S216 in Isomer 2, demonstrating EAD’s ability to differentiate positional isomers, a challenge for traditional collision-based MS/MS methods like CID.
Fast and flexible
EAD can be operated in data-dependent acquisition (DDA) or MRMHR mode, with a fast scanning rate (~20 Hz in DDA mode) and the ability to tune the electron kinetic energy (KE).
Powerful data analysis platform
Biologics Explorer software offers powerful, intuitive tools for peptide mapping and PTM analysis.
For methods, materials, and full results, read the complete technical note Comprehensive characterization of O-linked glycosylation in etanercept by electron activated dissociation (EAD).