Using transposases to generate clones with high, predictable expression and stability
Standard cell line development workflows include transfection followed by the screening of hundreds to thousands of clones, in an effort to find high producing clonal colonies that are stable and demonstrate specific attributes based on the product specifications. Some approaches at the clone isolation stage utilize in-situ fluorescence detection to try and pull out up front the best protein secretors.
I am pleased to share the following guest blog that presents an alternative solution to ensure isolation of the high producing clones – transposases. I was fortunate enough to be able to interview the author about his article and have provided the transcript of our conversation following the guest blog.
Elimination of fluorescence approaches at the clone isolation stage of cell line development: a paradigm shift
A Guest Blog by Dr Ian Taylor, Chief Commercial Officer, Solentim
Back in the early summer at ESACT in Copenhagen, I was impressed once again with the extremely high quality of the poster sessions throughout the event, presented by top level scientists working in labs around the world.
One aspect that surprised me however, and stimulated me for the theme of this blog, was the number of posters which described using fluorescence in their research to facilitate clone isolation for potentially the highest producing clones.
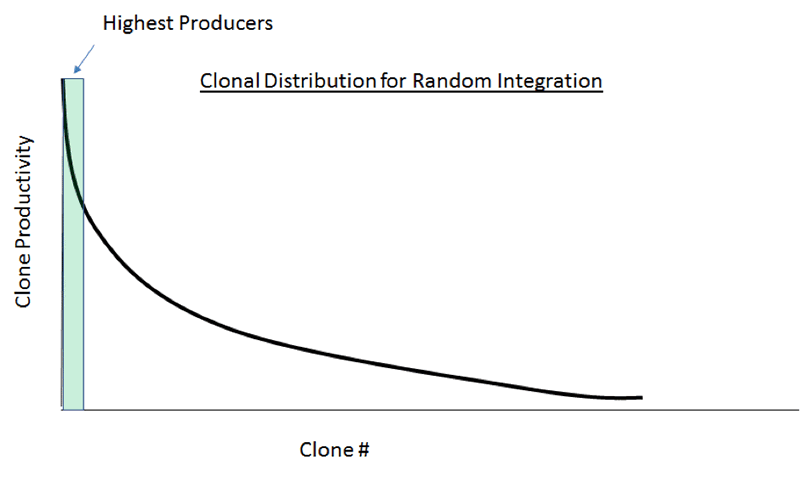
There is undeniably still a sense amongst a good number of those involved in cell line development that, following transfection, it is necessary to screen thousands of clones in order to identify the small subset which are the high producers. The principle is that using fluorescence detection, either with a labelled antibody or peptide, it is possible to detect, and subsequently isolate, the cells producing the highest amount of antibody (or any product) very early at the single cell stage. This method is based on a typical clonal distribution for random insertion cloning as shown below (Figure 1). On this graph, high producers represent only a small subset of the population, as denoted in the green box.
The concept of screening out “gold nugget” high producers with fluorescence is not new; whilst working at Genetix back in 2005, myself and colleagues developed the ClonePix FL for precisely this purpose. The ClonePix FL enabled cells to be isolated in semi-solid media, then as these grew and secreted antibody into the media, a fluorescently labelled secondary antibody was used to detect and “pick out” high producing clones.
However, it later became apparent that there was a massive flaw in this approach, and this same flaw exists for current methods using fluorescence detection. Namely, there is a lack of correlation between fluorescence at the single cell stage compared with the eventual productivity for expanded clones in shaking media. In reality, the fluorescence simply eliminates the population of non-producing clones (those not fluorescing in the screen) rather than identifying high producers. For producers, any ranking for productivity at the single cell isolation stage simply does not translate into productivity performance in culture.
In my opinion, it is now crucial to ‘bang the drum’ for a different approach which changes the dynamics of the cell line development workflow. This approach is to use transposases. By using clever vector design and targeted gene integration, it is possible to create a population whereby the majority of clones have the GOI inserted into specific sites, generating clones with high, predictable expression and stability. Some pharma companies, such as Pfizer, have their own proprietary approaches using vectors containing ‘landing pads’ to achieve this. However, transposases offer an excellent commercial alternative.
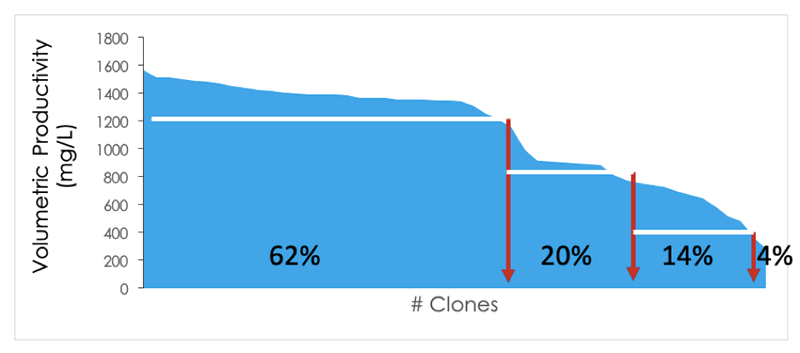
Solentim has recently partnered with ATUM– one such company providing transposases (Leap-in™). The game changer here is that transposases shift the clonal distribution massively towards high producing clones; for example, in Figure 2 below, 62% of clones are shown to be in the first quartile for productivity.
In practical terms, this entirely removes any rationale for using fluorescence for clone isolation, as the majority of clones from transfection will inherently be high producers. Therefore, the only requirement for successful cell line development is an efficient, reliable single cell isolation and documentation system (e.g. the VIPS). An additional benefit of this transposase approach is that clones also exhibit high inherent stability, therefore taking stability studies off the critical path (saving even more time).
Further endorsement of the impact of transposases has been demonstrated by Lonza’s GS piggyBac™ transposases.
The take-home message here for cell line development groups is to understand that fluorescence assays for trying to isolate the top clones are an outdated approach, providing a binary result of producer vs non-producer. Contrastingly, using transposases is quick, easy and only requires a handful of plates for a complete project. Find out more about the role of transposases and the VIPS in cell line development and biotherapeutics here. Please do get in touch with any questions about our products or transposases.
Interview With Dr Ian Taylor, Chief Commercial Officer, Solentim
Why do you think that fluorescence detection is still used in clone isolation when, as you describe, there are other technologies available?
I think it is partly caused by new groups coming into the field who are not familiar with the historical experiences of ClonePix and the practical workflow pitfalls with this approach.
Can you summarize what you see as the key advantages to transitioning to a transposase approach?
Simple, you can complete your whole cell line development project in a handful of plates, rather than tens of plates using random integration. Clones from this approach are also inherently very stable.
What do you think the time savings are when using a transposase approach vs. fluorescence detection?
Hard to say, as both should generate plates with clones which are predominantly all producing the protein. Typically, for a system based on fluorescence isolation of clones the downside would be more the very high initial capital investment costs and ongoing detection reagent costs. I should point out that fluorescence assays are still useful in the next stage after clone isolation, for measuring protein productivity in the cloning and expansion plates.
Can you tell us a little more about how the transposase approach can be coupled with VIPS in cell line development and also the benefits of this approach?
Transposases are not specifically coupled to VIPS, it is just our recommendation for the cloning and transfection stage. In terms of workflow benefits, the VIPS will ensure high seeding efficiency by giving a single cell isolated in most wells of the plate, and by virtue of the transposase approach, most of the single clones will be high producing clones. The result is that you can reach your project target number of high producing clones with only a handful of plates.
For more information, please see Cell Line Development Workflows