
mRNA-LNP integrity profiling – Ask the Expert
The use of lipid nanoparticles (LNPs) as non-viral delivery tools for nucleic acid payloads, such as messenger RNA (mRNA), has become more widely adopted in the industry not only due to its use in COVID-19 mRNA-based vaccine formulations. The transformative potential of gene therapy for modern medicine has driven many biotech and biopharma companies to invest in developing nucleic acid-based therapeutics for cancers, rare diseases and more, beyond the scope of prophylactic vaccines and LNPs are seemingly one of the most advanced technology for delivery.
The quality of encapsulated mRNA is critical to ensuring the efficacy and safety of the final vaccine or therapeutic. However, its large size and fragile nature pose challenges for analytical assays to assess mRNA quality attributes thoroughly. Furthermore, the demand to get to market faster is putting increasing pressure on companies. Turnkey workflows and higher throughput instrumentation can help reducing the timelines to get to valuable answers. However, quality of data and results should not suffer.
That is why SCIEX developed the BioPhase 8800 system, an eight capillary electrophoresis system, providing excellent resolution of nucleic acid species. With capillary gel electrophoresis with laser-induced fluorescence detection (CGE-LIF) intact mRNA species and degradation products can be analyzed to assess mRNA integrity and quality. The LIF technology provides highest sensitivity to avoid missing lower abundance species.
Assay development can be weeks’ to months’ worth of time, which can be significantly reduced, using the RNA 9000 Purity & Integrity kit, a kit that can be used for sizing and quality assessment of single-stranded nucleic acids using the PA 800 Plus or the BioPhase 8800 system. Since transferability of assays between instruments and departments can help reduce bottle necks during development all the way to quality control (QC), it is worth noting that developed workflows can be transferred between both CE platforms.
The CGE-LIF method provides an amplification-free workflow for effective characterization of RNA products and profiling of nucleic acid impurities.
We had the opportunity to speak with a SCIEX expert, Dr. Fang Wang, who is a Sr. Technical Product Manager, to answer some frequently asked questions about CGE-LIF for mRNA profiling and to provide our readers with practical tips for implementation of this workflow in their own labs.
Our questions and answers are below.
For LNP encapsulated mRNAs, what is the best sample preparation method to extract the mRNA for characterization? Does the lipid composition impact the results?
That’s a great question. We have done studies in collaboration with researchers at SINTEF and found that Triton X-100 treatment combined with heat and the addition of formamide was the most effective method to release the encapsulated mRNA and to avoid secondary structures prior to analysis with CGE-LIF. The final concentration of the Triton X-100 lysis solution used in our studies was 0.2% and samples were heated at 70 ⁰C for 10 minutes to extract the mRNA from mRNA-LNPs, followed by dilution with 80% formamide. It became evident that formamide is crucial to disrupt the formation of higher order structures and to increase the signal for the main species of mRNA.
We were able to use this extraction method to successfully evaluate mRNA size, integrity and nucleic acid degradation for different nucleic acid payload concentrations and different lipid compositions. The samples we analyzed contained different ionizable lipids, commonly known as MC3 and SM-102. MC3 was used for the formulation of the first FDA-approved siRNA therapeutic, while SM-102 is in use for the COVID-19 mRNA vaccine by Moderna.
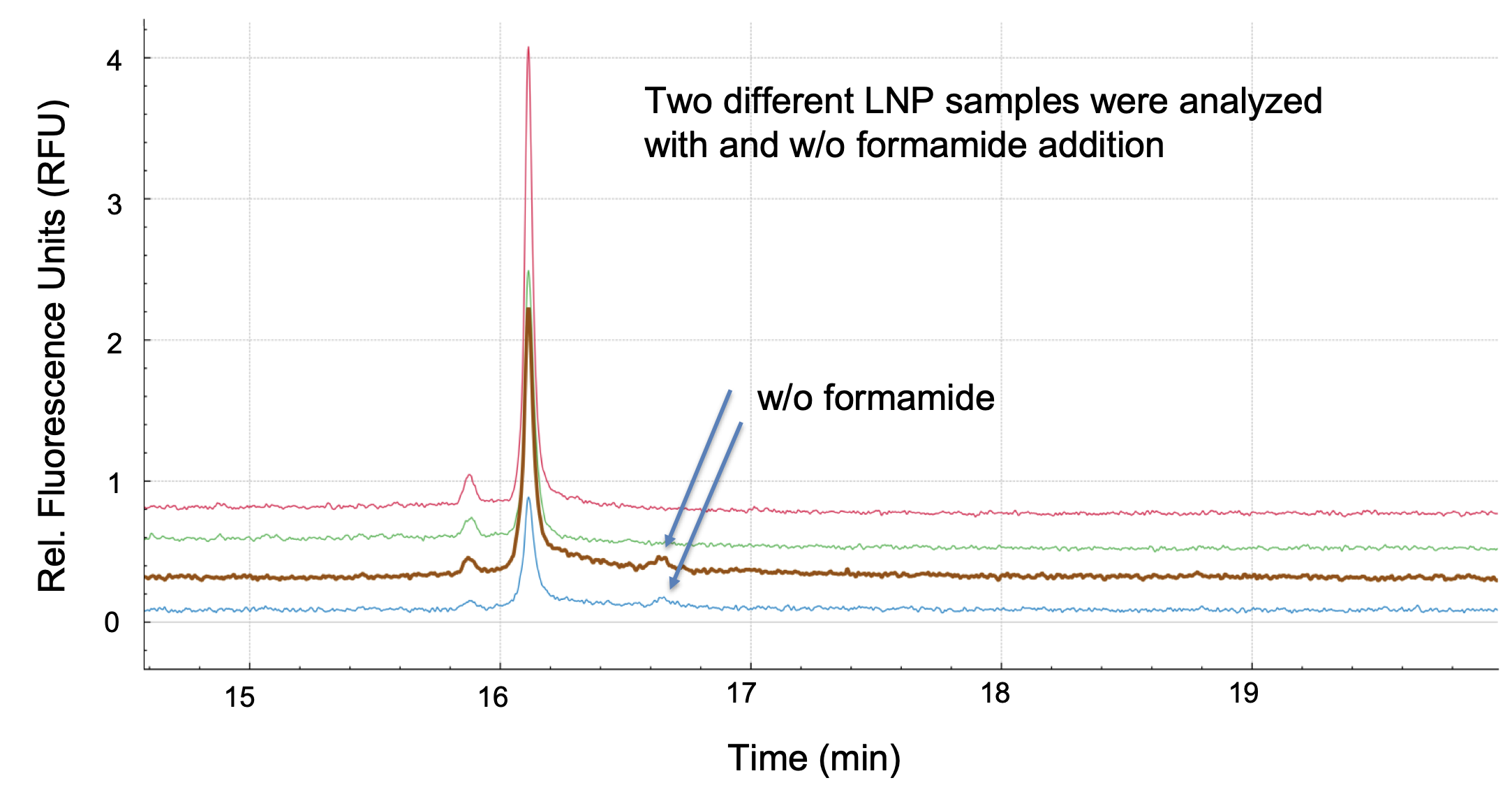
Since there is a wealth of different lipid compositions in use — especially proprietary ionizable lipids — we need to ensure that the extraction method works, and that the quantification is unaffected by differences in LNP composition. The preliminary results indicate the method has great platform potential.
How many different sample preparation procedures have you tested? How much better was Triton X-100 compared to other procedures?
We have tested more than 5 different sample preparations. So far, Triton X-100 offers the best results in terms of reproducibility, feasibility, and extraction of the mRNA from the LNPs without affecting the RNA measurements.
We heard that if lipids were not removed from the mRNA-LNP complexes prior to CE analysis, then the capillary life was decreased. Is that a common observation?
Based on our experience, the lipids do not negatively affect the CE measurement. We do not remove the lipids after sample preparation prior to injection onto the CE system as this step bears a high risk of affecting the mRNA and related quantification. If decreased capillary lifetime is observed, I recommend investigating other components that might have been introduced during sample preparation, such as other detergents. Those could potentially damage the capillary. Users should therefore exercise caution with the type of solvents or solutions applied for the RNA extraction from mRNA-LNP, and with subsequent injections of these solvents into CE instruments in general.
What is the sensitivity of detection using this method for mRNA profiling?
We observed great sensitivity with a lower limit of detection (LLOD) of 6.10×10-6 mg/mL and a lower limit of quantification (LLOQ) of 1.22×10-5 mg/mL. The linearity was excellent with an R2 value of 0.9999.
Throughput is of high importance to many labs. How long does it take to run 8 samples, including sample preparation and analysis?
The sample preparation time takes around 15 minutes, which includes the time it takes to mix reagents, heat for 10 minutes and cool down the sample. For mRNA-LNP samples in particular, a lysis buffer solution containing Triton X-100 is added for 20 minutes prior to utilizing the heat denaturation step. Setting up the reagent plates for the CE system will vary from 5 to 10 minutes. The length of the run time for eight samples depends on the platform — the multi-capillary BioPhase 8800 system or the single-capillary PA 800 Plus system. When using the BioPhase 8800 system, the run time is about 25 minutes per 8 samples on the BioPhase 8800 system or 25 min per 1 sample on the PA 800 Plus system.
Has the RNA 9000 Purity & Integrity kit been used to qualify mRNA therapeutic modalities besides mRNA-LNP?
Yes, the RNA 9000 Purity & Integrity Kit has been used to characterize the CRISPR/Cas9 gene editing system. The kit was used to analyze the main product of the Cas9 mRNA and the single-guide RNA (sgRNA) component of this gene editing system along with nucleic acid impurities on both the PA 800 Plus system and the multi-capillary BioPhase 8800 system. The ladder included in the kit ranges from 50–9,000 bases, which enables the analysis of Cas9 mRNA and sgRNA at the same time.
Can the RNA 9000 Purity & Integrity kit be used for DNA-based products?
Yes, the kit can resolve single-stranded (ss) nucleic acid products in the range of 50–9,000 bases within approximately 25 minutes. That includes ssDNA and ssRNA applications. Scientists and QC personnel can take advantage of this assay in R&D and QC by using the high-throughput BioPhase 8800 system or existing PA 800 Plus systems.
From your unique perspective, how do you think this technology will impact the development of new nanomedicines?
The ability to do this type of high-resolution analysis can help the field in the development of new mRNA-LNP products by enabling better understanding of how changes in the LNP composition or varying storage conditions can affect the mRNA’s integrity profile. Being able to quantify the quality and stability of the mRNA-LNP product, assess nucleic acid fragmentation and impurities with high quality data in a high throughput manner can provide scientists with information that accelerates the selection of the best LNP formulations for vaccine and gene therapy development. Optimal RNA extraction, separation, detection, and quantification using the RNA 9000 Purity & Integrity kit with CGE-LIF technology on the PA 800 Plus and multi-capillary BioPhase 8800 system along with automated software enables robust, high-throughput RNA stability and nucleic acid degradation profiling of large sample collections to support a better understanding of emerging mRNA-LNP modalities.
For more information, please see RNA Analysis Resource Center