Single-Step Cloning Strategy Speeds Timeline for Novel Vaccine Candidates
With the emergence of Covid-19 this year and the threat of other novel viruses looming on the horizon, now more than ever we need fast, efficient methods for generating novel vaccine candidates. These new methods could be developed as new technologies, rethinking existing technologies, or combining existing technologies in a new way. I am pleased to share the following guest blog, which presents a method developed by GSK for single-step cloning to accelerate the novel vaccine development timeline. The approach combines existing technologies to shorten the cloning process for CHO cells to a single-step.
I was fortunate to be able to interview the author about his article and have provided the transcript of our conversation following the guest blog.
GSK pioneer a single-step cloning strategy to generate novel recombinant sub unit vaccine candidates.
A Guest Blog by Mark Stockdale, Solentim
GSK Vaccines’ team demonstrate the benefits of a new, single-step cloning strategy to generate CHO cell lines expressing novel vaccine candidates, reducing effort and shortening timelines.
Currently we are living in unprecedented times with a global health threat from a novel, highly infectious respiratory disease. We are trying to prevent a large percentage of the population succumbing to this virus through early detection and isolation strategies. There is the potential need for a long-term treatment to reduce fatality rates if variants of COVID-19 persist in the human population. New vaccines and more rapid development timelines are needed to provide immunity to this new threat, and others like it.
A fantastic effort has started, with scientific researchers and vaccine manufacturers working hard to abate this crisis; close to 150 COVID-19 vaccine candidates are now listed by the WHO1 ranging from novel RNA-based vaccines to the re-purposing of already marketed drugs. A successful drug could curb transmission and reduce the respiratory burden on those infected with COVID-19. One route to vaccine discovery is to analyse the antibody repertoire of people who have recovered from the infection, use B-cell screening to find an active antibody, or generate protein subunits to prime the immune system. In order to effectively scale therapies of this nature, we will require a fast and effective way to create cell lines producing the recombinant subunit vaccine protein(s), which shows appropriate antigenicity and high yield.
GlaxoSmithKline (GSK) has a strong track record in developing vaccines for some of the most prominent threats to humankind. Recently Xiangming Li2 in Marcin Bugno’s team at GSK Rockville looked at their capabilities to generate multi-subunit recombinant vaccine proteins against human CMV (cytomegalovirus), and the technology that could help reduce the response time and effort required.
Assessment of the classic cell line development process using CHO hosts as the cell factory shows that the analysis of clones is limited in the early stages, focusing instead on clone outgrowth to determine performance. In part, this is due to the lack of sensitive assays for productivity assessment and accurate methods to determine cell number, allowing specific productivity to be determined, a strong predictor for lead clones.
Li et al2 comment that, although new instruments have reached the market in the last two years, these systems are focussed predominately on classical antibody workflows, with assays limited for productivity assessment of monoclonal IgG antibodies in a semi-quantitative manner: ‘Beacon (Berkeley Lights), utilizes nanofluidics and OptoElectro position technology to achieve single-cell cloning and high cell density culture conditions, potentially providing an advantage for selecting clones compatible with upstream bioreactor processes. However, similarly to ClonePix technology, the productivity is estimated indirectly, based on diffusion-based staining of the secreted antibody with a fluorophore-tagged small molecule binding human IgG Fc’.
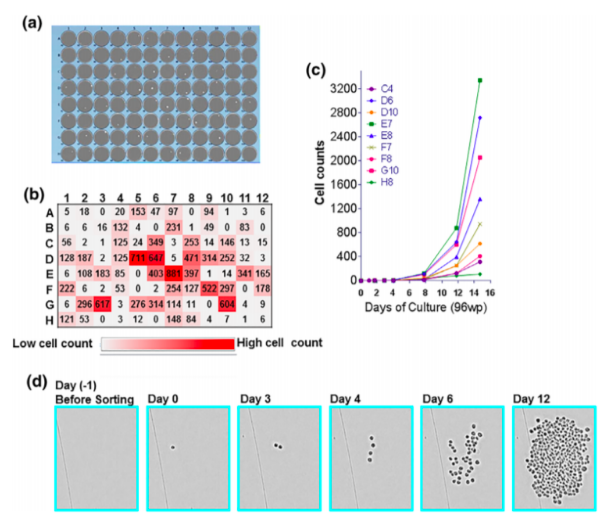
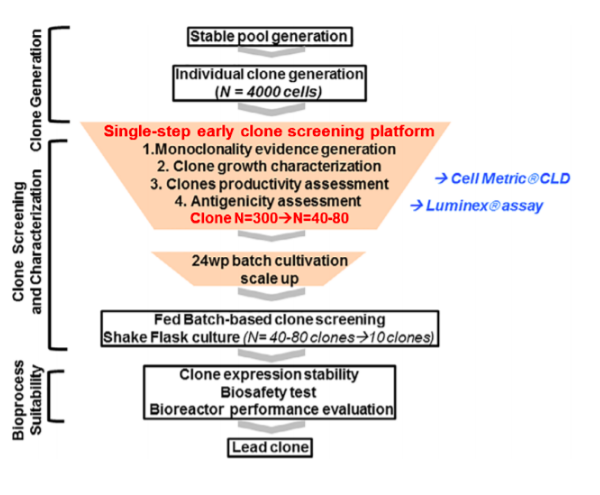
With the expanding need to express antibody fragments and novel proteins, this calls for flexible and multifaceted workflows which leverage the capabilities of manufacturing groups and discovery groups to assess the performance of the cell lines during primary screening. The workflow devised by the team at GSK uses traditional FACS to deposit single cells into 96-well multi-well plates. The presence of a single cell in each well is confirmed by the Solentim Cell Metric® CLD, according to regulatory recommendations, to confirm a cell line is derived from a single cell.
The growth of each single cell was then monitored using the Cell Metric CLD to accurately determine the cell number per well in a 10-14 day window (see figure 1). The outgrowth reported from single cells was approximately 20%, which is typical for the CHO platform used in the experiment. At day 14, half of the supernatant was harvested from the single-cell isolation plates for quantification of the multi-subunit protein. The volumetric titre was anticipated to be in the ng/mL range, far below the sensitivity of many high-throughput quantitative assays, so this required the adoption of a multiplex immunoassay with sensitivity in the ng/mL range.
Can you describe a standard cloning workflow and timeline?
The cloning workflow has dramatically changed over the last two decades and the health regulators have been keen to keep pace with this. Many labs will have slightly different workflows based on the technology they have at hand so ‘standard’ is hard to define. One thing I can be sure of; the classical use of manual limiting dilution has been perceived as a low barrier of entry to perform single-cell cloning, but it will cost a company time and money in the long term.
In a classical workflow – such as the one described above – two rounds of limiting dilution will be performed based on position distribution, with many wells remaining empty to provide a high probability that the cell line is derived from a clone. A CLD process that took 6 months can be brought down to as little as 4 months if the need for limiting dilution can be removed. There is also an additional cost from the complexity of handling the clones, wasting media and plates to obtain a degree of certainty in plating a single cell in a proportion of the wells in a microtiter plate. Timescales will also vary based on other factors like the cell line being used.
The decision for when a company switches from manual to instrument-aided single-cell cloning can be a complex one based on the size of the cell line development pipeline and the intended exit strategy. For me, it is an inevitable adoption for cell line development laboratories where such high value is still placed on clonal derivation of cell lines. The deposition of a single cell into the well is a whole other story.
With respect to the work GSK has done to develop a single-step cloning strategy, can you describe the impact that implementation of this process had on the cloning timeline and overall process development timeline?
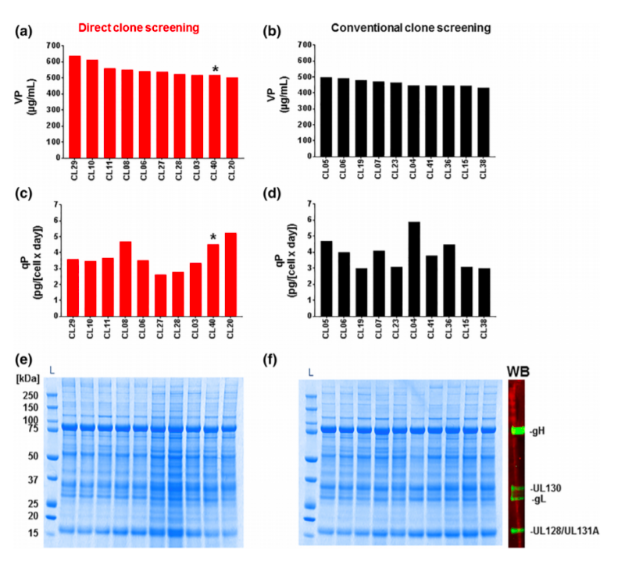
The authors refer to an overall saving on time by implementing the single-step cloning strategy versus the iterative traditional process, but within the paper they do not put a number to the time saving. By implementing an early screening strategy the authors describe a rapid reduction in input number from 4000 clones down to 40 – 80 top clones by using the combination of the Cell Metric and Luminex assay. In the traditional process, they talked about selecting 300 clones at the same stage, so this is a significant reduction in clone handling and complexity in the lab. I would much prefer to passage 40 cell lines rather than 300!
In the GSK study, the Solentim Cell Metric CLD was used to ensure the cell line was derived from a single cell. Please describe what this workflow looks like compared to standard cloning steps.
By bringing a dedicated imaging system into the process, we can now switch the conversation from the probability of clonality to assurance that the cell line was derived from a single-cell clone. Health authorities are becoming more familiar with this data, and the likes of the FDA have indicated that this is their preference going forward.
In the study, the team adapted a Luminex bead-based sandwich immunoassay to speed throughput, would you elaborate on how the Luminex assay worked in sync with the Solentim Cell Metric CLD to achieve single-step cloning?
The major benefit of the Luminex assay was the ability of multiplex assays to detect the five subunits of the product they were producing, to determine antigen quality and generate an assay that was sensitive down to the ng/mL range to determine volumetric titer from a small number of cells in the colony, around 500 cells.
With the high-throughput and high-resolution imaging capabilities of the Cell Metric CLD, the team was able to determine cell counts and growth patterns of individual clones during outgrowth in the single-cell cloning step. By combining the sensitive volumetric titer values from the Luminex and the cell counts from the Cell Metric CLD, the team could rank the clones based on specific productivity (pg/day/mL) and antigen quality. Comparatively, the traditional method was a somewhat random selection with little correlation between top clones in low volume culture at clone selection and the scaled-up culture.
Lastly, how could this technology combination workflow be applied to other processes?
The example from GSK was for a multimeric protein being used as a vaccine. The challenges with these complex molecules are how they fold together and form the viral protein antigen complex which primes the immune system to recognise the virus. I can see the same difficulties with novel biotherapeutics built around Fab regions of IgG antibodies. These chains are being constructed into complex structures to bind to multiple antigens, often termed bispecific antibodies. The folding of these multichain Fab containing molecules can result in misfolding or daisy-chain like structure leading to a low proportion of the correct and active form of the molecule. By using functional screens early in the process, we can focus on the cell lines producing the most viable molecule to take forward to the clinic. The challenge to create these assays is finding a ubiquitous assay that recognises the majority of molecules progressing through the cell line development pipeline, rather than building an assay for each molecule.
Visit the Solentim website to find out more about the Cell Metric® and how it helps to accelerate cell line development for vaccines research.
About the Interviewee:

Mark Stockdale, Solentim
Mark Stockdale is a keen technology pathfinder and works within Solentim to amalgamate the technical and business functions within the company. Prior to Solentim, he worked as a Group Leader in Horizon Discovery Ltd, a scientist within Lonza Biologics, Cyclacel Ltd and started his career at Pfizer. With over 15 years’ professional experience in cell line development and drug discovery, his passion is to bring new technologies to bear on unmet needs, he provides great insight into the future technology requirements in stable cell line development.